Profile fresh, frozen, and fixed samples—even FFPE—with sample prep methods optimized for your success. With 40+ Demonstrated Protocols across the breadth of our portfolio, your single cell studies can go further than ever before.


- Capture data from a wide range of species with our reverse transcription (RT)-based Universal assay
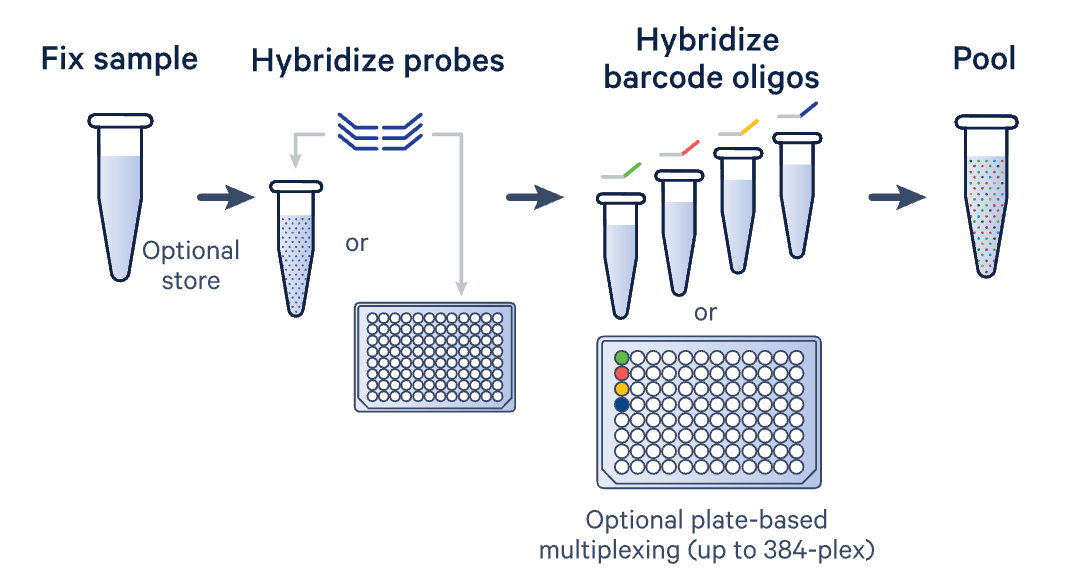
- Expand the scope of your studies to include FFPE samples with our probe-based Flex assays
- Easily isolate nuclei from frozen samples with kitted reagents and a unified workflow
- Easily move from pilot studies to ambitious experiments with the ability to process 1–3,072 samples, and profile hundreds to millions of cells per run
- Batch and run assays on your schedule with robust fixation protocols, even for whole blood samples
- Reduce reagent volumes and costs with easy on-chip multiplexing for Universal 3' and 5' assays
- Use plate-based multiplexing for fixed samples to profile up to 8M cells per run and enable sample batching


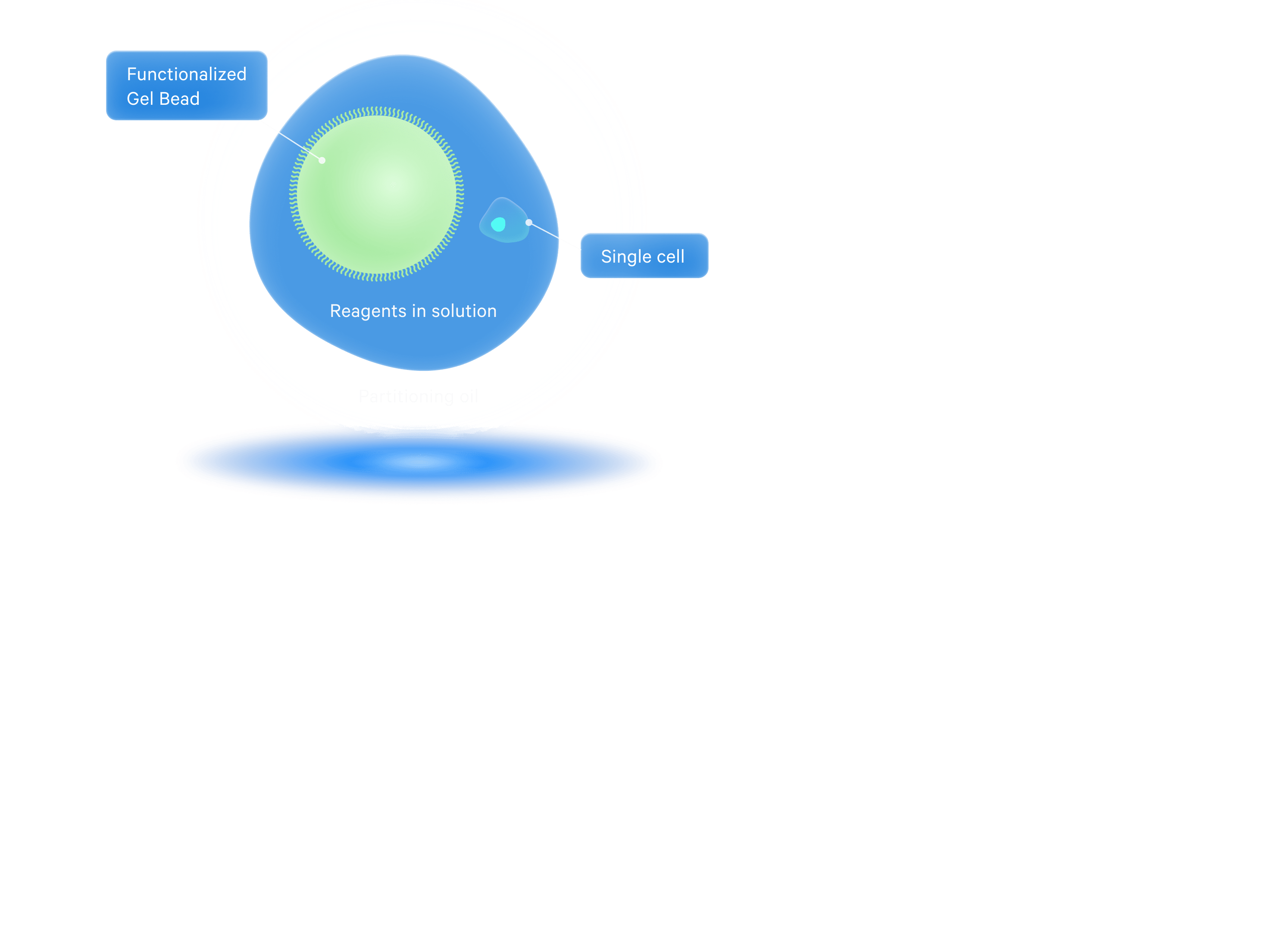
Within Chromium X series instruments, cells move through the channels at a limiting dilution, generating 10,000+ nanometer-sized Gel Beads-in-emulsion (GEMs) in 4 minutes.
Each GEM is the site of a micro-reaction, in which Gel Beads are dissolved and target molecules from each cell are captured and barcoded, marking each target molecule's cell of origin.
Did you know? Chromium X series instruments automate the most critical steps in single cell assays: partitioning and barcoding.
These fast, efficient workflows reduce technical errors and batch effects common in labor-intensive, time-consuming manual workflows. Plus, the instruments combined with GEM-X advancements deliver industry-leading cell capture of up to 80% and superior reproducibility.

Go from sample to your sequencing-ready library in one day, with just 3 to 4 hours of hands-on time.
Barcoded fragments from GEMs are pooled for downstream reactions to create Illumina sequencer-compatible libraries:
- MiSeq
- NextSeq 500/550/2000
- HiSeq 2500 (Rapid Run)
- HiSeq 3000/4000
- NovaSeq 6000


Our intuitive and powerful software was developed to accelerate your access to experimental conclusions, whether you’re a seasoned pro or looking at your first dataset. Identify cell types and states, detect rare transcripts, perform multi-sample comparisons and more—all with free, user-friendly software.

- Unlock downstream analysis with Cell Ranger, our easy-to-use pipelines that set the standard for scRNA-seq data processing
- Get results quickly and run Cell Ranger pipelines on 10x Genomics Cloud Analysis, our optimized cloud infrastructure
- Visualize your processed data with the interactive Loupe Browser, quickly plotting differential expression of your favorite genes
- Take advantage of continued innovations for new features like automated cell annotation

- Three-fold coverage of targets for high sensitivity and exceptional performance for FFPE and low-quality samples
- Run large-scale studies (up to 384 samples a week) with convenient, plate-based multiplexing

- Gather broad information (isoforms, single nucleotide polymorphisms, long non-coding RNAs, etc.) from diverse species
- Bring easy multiplexing to samples that can't be tagged (e.g., non-human/mouse, or nuclei) or samples that are limited
- Batch and multiplex to enable your largest studies, processing up to 384 samples or up to 100M cells per week
- Scale at your pace by running partial plates, using only the reagents you need, when you need them
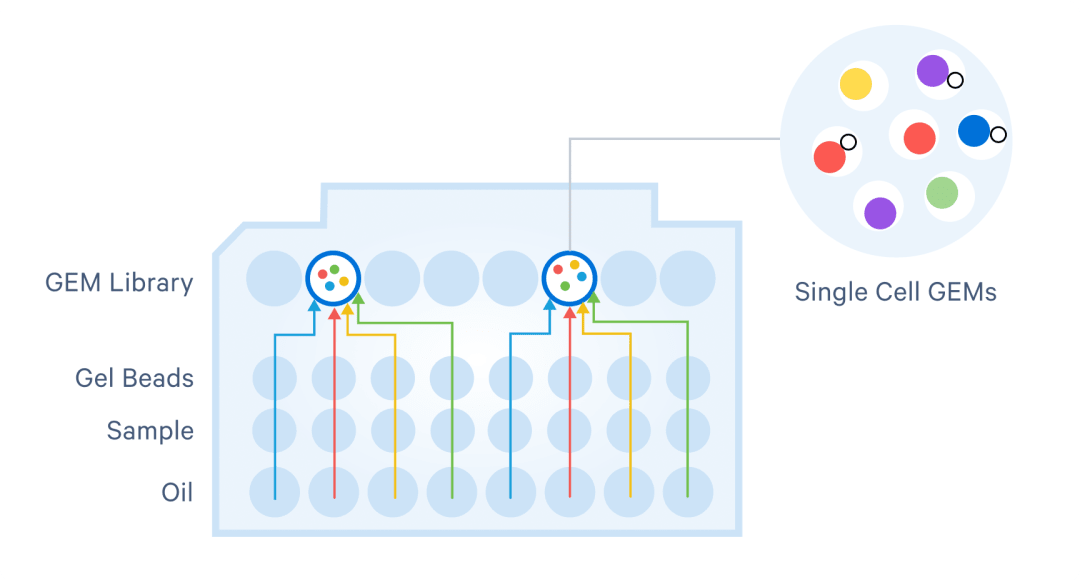
- Process 1–8 samples and up to 40K cells/chip , with on-chip multiplexing for for any study, even at small scale
- Add multiplexing to your workflow easily without the need to tag samples upstream of chip loading
Platform |  Chromium Single Cell |  Visium Spatial |  Xenium In Situ |
|---|---|---|---|
When to use | Comprehensive single cell data Ideal for deep characterization of cell populations and states. | High-resolution spatial gene expression Understand complex tissues, neighborhoods, and cell to cell interactions. Integration with other spatial-omics, histology, and morphology. | |
Why to use | Unbiased single cell discovery High per-gene sensitivity | Unbiased spatial discovery | Targeted spatial exploration High per-gene sensitivity |
Applications | Whole transcriptome gene expression Protein TCR, BCR CRISPR ATAC | Whole transcriptome gene expression | Targeted gene expression (up to 5,000 genes) |
Resolution | Single cell | Transcripts assigned to 2-µm areas | Single cell |
Data readout | NGS-based | NGS-based | Imaging-based |
Sample compatibility | Single cell or nuclei suspensions from fresh, frozen, or FFPE samples | FFPE Fresh frozen Fixed frozen | Fresh frozen FFPE |