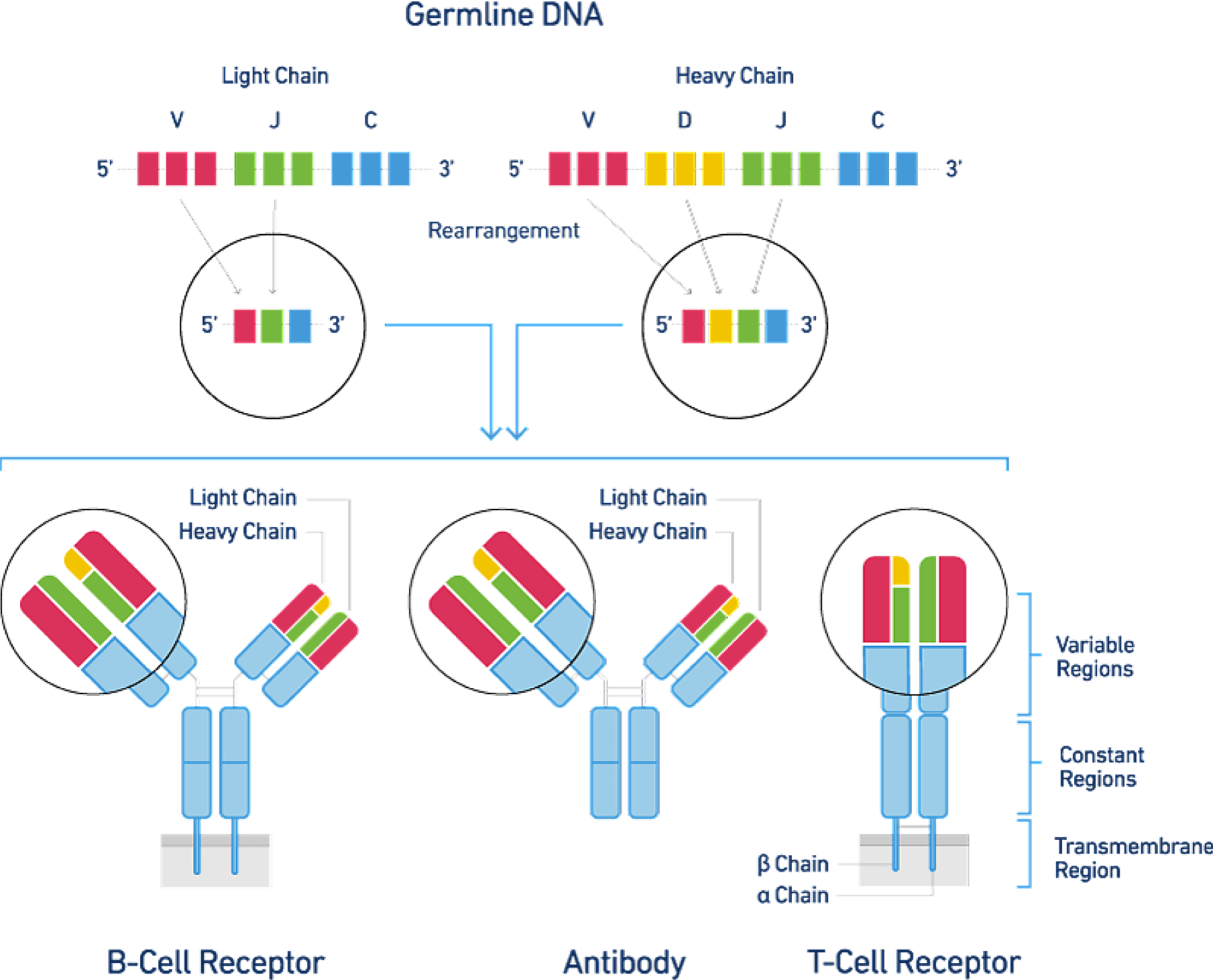
Are you truly tracking your cells’ transcriptomes, or are you just reading into the averages?
Single Cell Resolution
Traditional RNA-seq methods analyzed the RNA of an entire population of cells, but only yielded a bulk average of the measurement instead of representing each individual cell’s transcriptome. By analyzing the transcriptome of a single cell at a time, the heterogeneity of a sample is captured and resolved to the fundamental unit of living organisms—the cell.
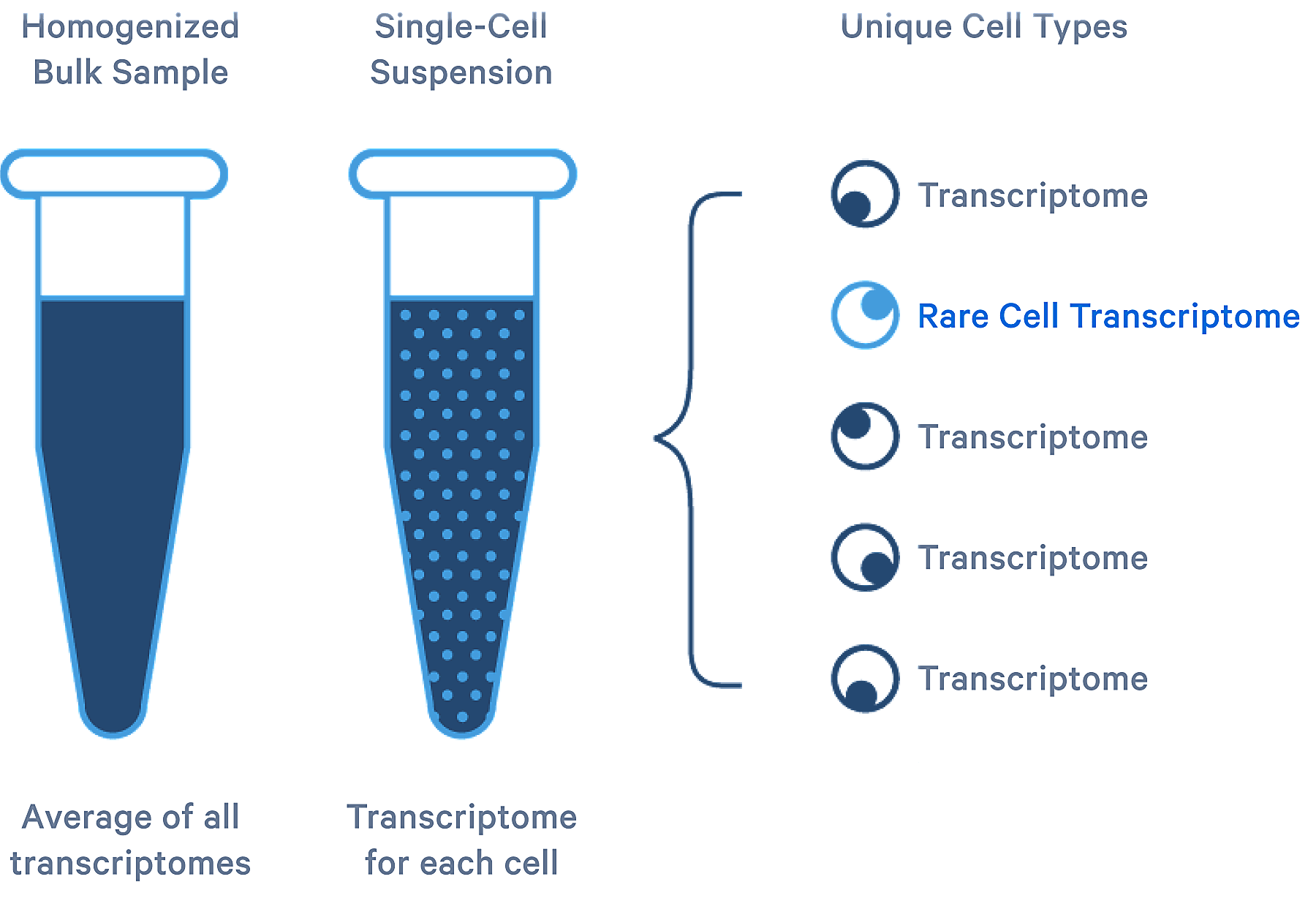
Tracking the trajectory of cell fates
Whether you’re working with tumor cells, stem cells,T-cells, or embryonic cells, heterogeneity is ever-present. scRNA-seq allows you to analyze the complexity of biological systems, both within a population and over time, at the single cell level. Single cell transcriptome analysis has enabled a detailed and unbiased look at this dynamic process in all its forms.
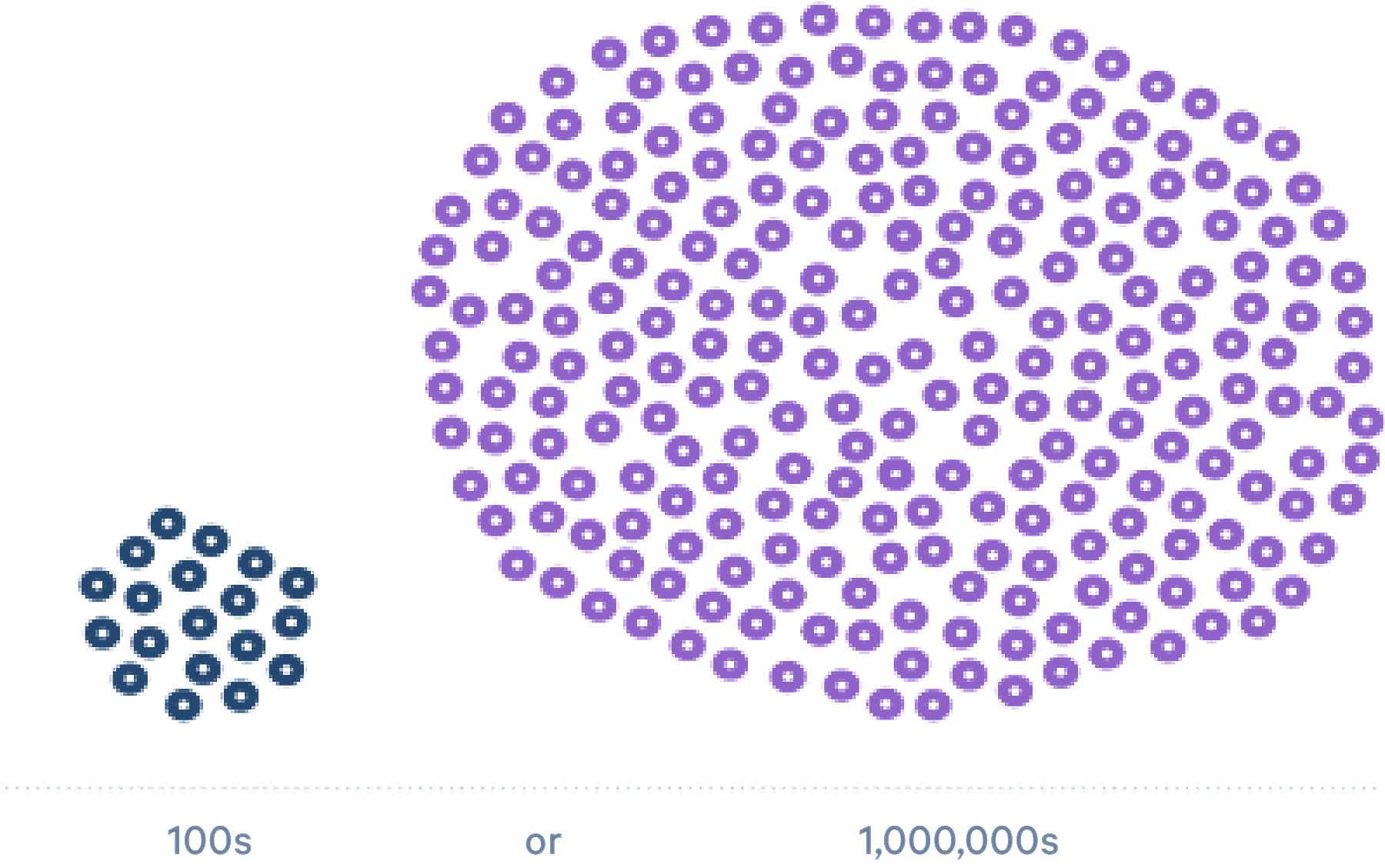
Scale 100s to 1,000,000s
Should there be an upper or lower limit to the number of single cells that must be characterized for meaningful analyses? The newest technologies for scRNA-seq enable you to catalog the cellular heterogeneity in your tissue, whether you have a scarce, rare sample of very specific cells or an abundant sample with many cell types. Droplet-based scRNA-seq methods with the highest capture efficiencies ensure that you see the whole picture.
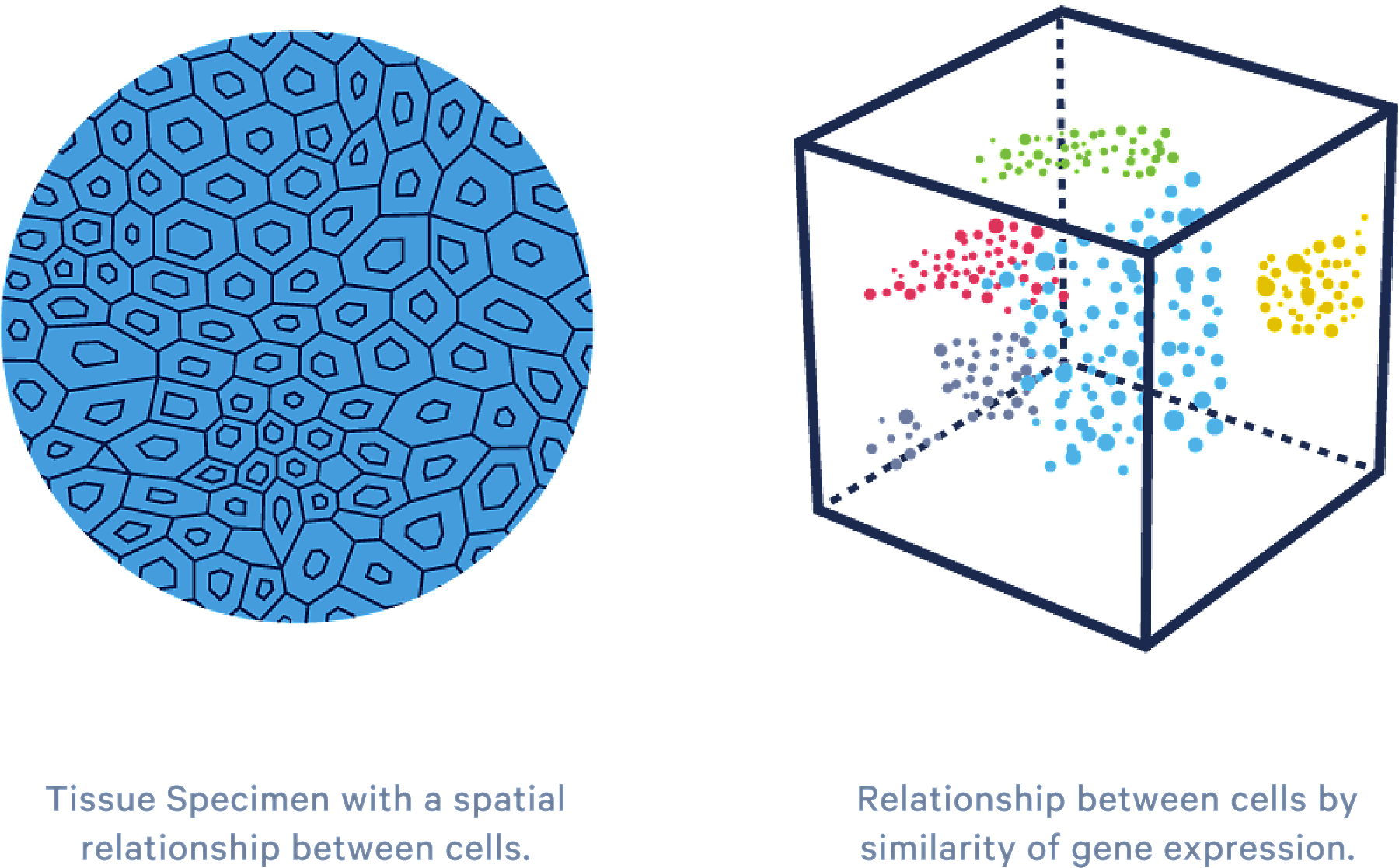
Clarify complex systems
For the first time, scRNA-seq is enabling a cell-by-cell molecular and cellular characterization of hundreds of thousands of cells within the same sample. Complex systems, like those found in the immune system, can be explored without limits. scRNA-seq can now be applied to:
- Immunology
- Neurology
- Stem Cell Biology
- Oncology
- Immuno-Oncology
- Functional Genomics