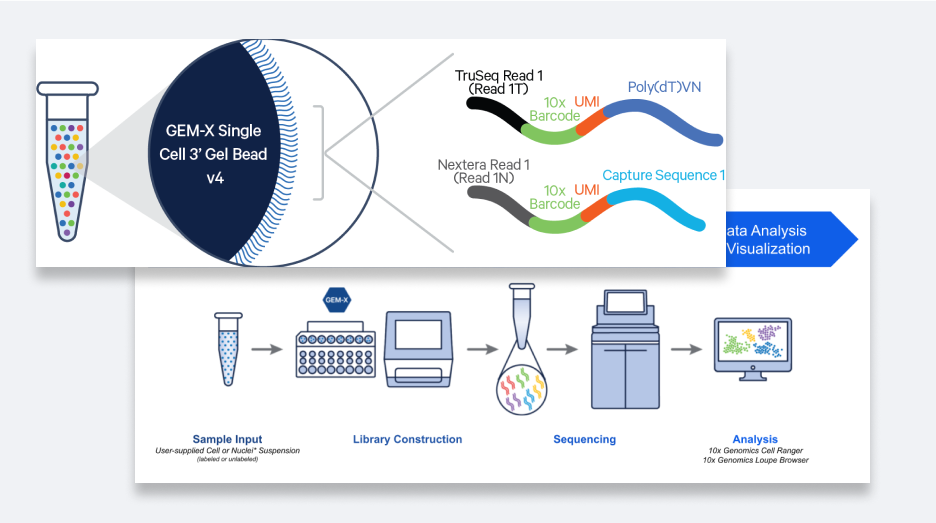
Single cell analysis enables meaningful discoveries
Bulk sequencing averages readouts, missing critical details driving complex biology.
Single cell sequencing allows scientists to see the unique gene expression patterns of each cell. This view more fully characterizes tissue heterogeneity, revealing the rare cell types that have big consequences in health and disease.

New to single cell RNA-seq?
The Chromium Advantage
- Higher gene sensitivity and up to 80% cell recovery efficiency.
- Enhanced detection of challenging cell types, including neutrophils.
- Reduce hands-on time and lower the risk of errors with instrument-supported automation of the most critical workflow steps.
- Produce higher quality libraries that can sequenced at lower depths.
- Backed by 10 years, over 2,200 patents, and >$1.5B in R&D investment.
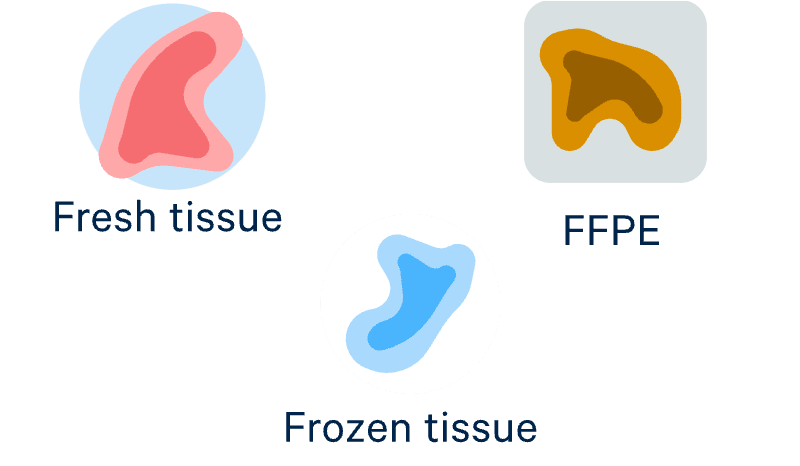
Access more sample types
Profile fresh, frozen, or fixed samples—even FFPE—with optimized protocols for proven performance

Automate critical steps to reduce error
Produce up to 2.56M barcoded partitions in just minutes, with up to 80% cell recovery
Construct high-quality sequence-ready libraries
Generate libraries with up to 95% usable reads, letting you detect more genes at lower sequencing costs

Analyze & discover with ease
Process and visualize your data with powerful software tools—no bioinformatics experiences required




Universal Gene Expression
Species agnostic. Maximum versatility.
Whole transcriptome coverage
Delivers broadest set of information, including isoforms, SNPs, etc.
Multiomic readouts from the same cell
3' or 5' gene expression
TCR/BCR
Protein
CRISPR
For cost-effective studies
Up to 160,000 cells (1-8 samples) per run, plus lower cost of <$600/sample
Flex Gene Expression
Fix, batch, and run on your schedule.
Protein coding gene coverage
Performs well with low-quality and FFPE samples
Multiomic readouts from the same cell
Gene expression
Protein
CRISPR
Highest throughput option
Up to 2.56M cells (1-16 samples) per run, for less than $0.01/cell
Products
Epi Chromatin
Unmask epigenomic profiles.
ATAC-seq chemistry
Explore open chromatin regions
Link directly to 3' gene expression (Multiome kit)
Multiomic readouts from the same cell
Chromatin accessibility
3' gene expression
Scalable nuclei preparation
Profile up to 80,000 nuclei (1-8 samples) per run
Products
David Michonneau, PhD
Professor of Hematology at Saint-Louis Hospital

Gene expression
Characterize the unique transcriptomic profiles of each cell in your sample


CRISPR screening
Directly link CRISPR guide RNAs to the resulting perturbed phenotypes
Practical considerations and best practices for getting started
Platform |  Chromium Single Cell |  Visium Spatial |  Xenium In Situ |
|---|---|---|---|
When to use | Comprehensive single cell data Ideal for deep characterization of cell populations and states. | High-resolution spatial gene expression Understand complex tissues, neighborhoods, and cell to cell interactions. Integration with other spatial-omics, histology, and morphology. | |
Why to use | Unbiased single cell discovery High per-gene sensitivity | Unbiased spatial discovery | Targeted spatial exploration High per-gene sensitivity |
Applications | Whole transcriptome gene expression Protein TCR, BCR CRISPR ATAC | Whole transcriptome gene expression | Targeted gene expression (up to 5,000 genes) |
Resolution | Single cell | Transcripts assigned to 2-µm areas | Single cell |
Data readout | NGS-based | NGS-based | Imaging-based |
Sample compatibility | Single cell or nuclei suspensions from fresh, frozen, or FFPE samples | FFPE Fresh frozen Fixed frozen | Fresh frozen FFPE |