This page covers frequently asked questions about standard and advanced workflows for Xenium panel design. Additional Q&A is available in Xenium panel design Knowledge base articles.
1. I began my panel design request without a design ID, and now I want to provide a design ID. Is this supported?
No, currently you must choose whether or not you will provide a preexisting design ID at the beginning of the panel design workflow. If you begin the panel design process without a design ID and later decide to provide an existing design ID, start a new request for that existing design ID and copy over your gene list and references from the initial design.
1. What are the input requirements for creating a Xenium custom panel?
Learn more on the Xenium Panel Design workflow guide page.
You will need the following for all Xenium custom panels:
- A gene list that contains the genes you wish to target in your Xenium experiment.
- A single cell reference dataset (accepted formats described here).
- The panel design tool provides a selection of curated references from CELLxGENE and GEO that can be used if they work with your experimental model.
- The references should closely resemble the tissue and condition of the samples you plan to use in your Xenium experiment.
- The references can be data from either 10x Chromium (Single Cell 3' Gene Expression or Single Cell Gene Expression Flex) or Visium (with reference-free deconvolution) platforms.
2. What happens when I reduce the number of probes per target?
We observe a roughly linear relationship between the number of probes for a target and the relative sensitivity of detection. This relationship is only true on the aggregate; individual probe behaviors will deviate depending on probe binding dynamics.
3. I am comparing data across multiple Xenium panels which have different probe set counts for overlapping genes. Do you have recommendations for how I should compare data across panels?
If the goal is to generate data for a specific gene that is directly comparable between different panels, consider whether the same probe sets are being used across panels for that gene. Due to our in silico screen for probe-probe interactions, these are not guaranteed to be the same across any two panels. However, this information can be found by checking the panel BED files, which are available as outputs of the custom panel design process and on the support site for pre-designed panels.
We have observed that sensitivity scales linearly, on average, with the number of probe sets in the range of 1-8 probe sets. If the panels have a different number of probe sets for the gene of interest, you can apply a gene-specific scaling factor to compensate for the sensitivity difference.
4. How do I check if there is a significant overlap between my genes of interest for a custom panel and Xenium pre-designed panels?
When you are requesting your custom panel in the Xenium Panel Designer app, we provide a warning on the panel design summary file if > 50% of your genes of interest are already included in any of our pre-designed panels.
5. Can a custom add-on panel designed for a particular Xenium pre-designed panel be used with other Xenium pre-designed panel products?
No, each custom add-on panel is designed for use with a specific pre-designed panel.
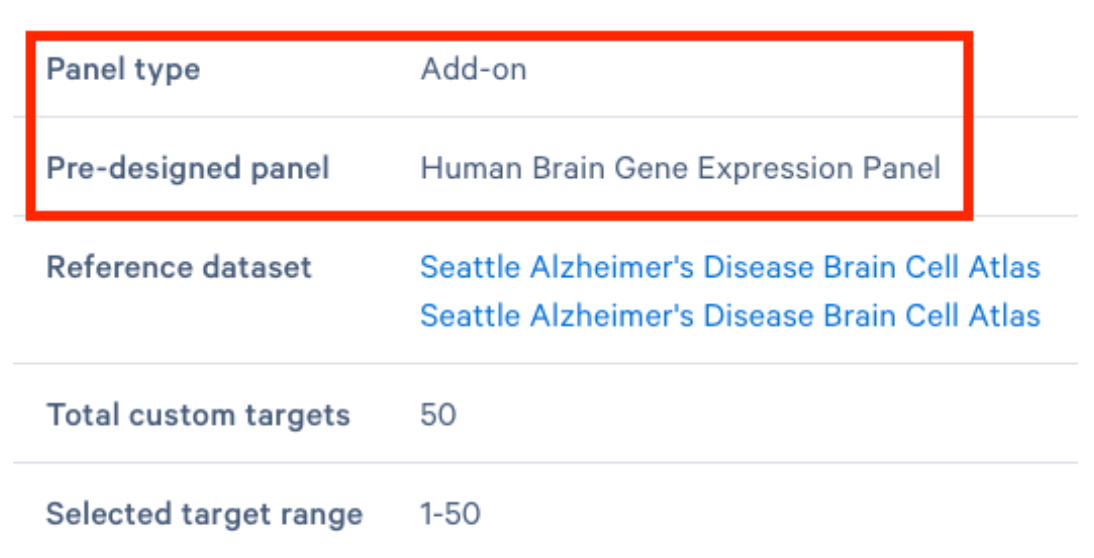
The "Panel Information" section of the finalized panel view in the Xenium Panel Designer app shows the pre-designed panel your add-on panel was designed for. For example, the screenshot below shows an add-on panel designed for the pre-designed Human Brain Gene Expression Panel.
6. Can I change or edit my design after finalizing the panel?
No, you cannot change or edit the design after finalizing the panel design request. If you want to request a similar panel to the one you have finalized, you can download the gene list CSV file from your finalized panel and use it as a starting point for the new request.
1. Why do I need a single cell reference dataset?
The single cell reference datasets are used to determine how much of the detection budget the panel is expected to use in the sample. This helps guide the selection of genes that can or cannot be successfully assayed, and provides a basis for deciding which genes require probe set coverage decreases. It allows the design tool to place coexpressed genes on optically distant barcodes to make the most efficient use of the optical space.
It is particularly important to find the best possible single cell dataset that you can (i.e., most similar to the sample you plan to use in your experiment) when working in diseased tissue. Learn more here.
2. What are the requirements for uploading my own single cell reference to the Xenium Panel Designer?
Your single cell reference must be unnormalized whole transcriptome feature-barcode matrices with cell type annotations for each matrix. The matrix and annotations should be bundled as a .zip, .tar, or .tar.gz file (one matrix + one annotation file per bundle). The accepted feature-barcode matrix formats are: 10x HDF5 or MEX. The 10x CLOUPE file format is also supported. Learn more about single cell references here and reference format conversion here.
3. What if I do not have a single cell reference?
We recommend that you have a closely matched single cell RNA-seq reference to accurately determine how much of the detection budget your panel is expected to use. It is particularly important to find a close match when working with diseased tissue types.
If you do not have an annotated single cell RNA-seq, you can use publicly available data from sources such as CELLxGENE. The Xenium Panel Designer provides a variety of curated reference datasets for both human and mouse tissues and a variety of conditions (see table).
1. What is advanced custom panel design?
Standard custom panels target well-annotated genes in the human and mouse 2020-A reference transcriptomes.
Advanced custom panel designs target sequences that we do not have existing probe designs for. Potential advanced custom applications include isoforms, gene fusions, viral or bacterial sequences, protein tags, fluorescent reporters, and transgenes. More information is available in the Choose Panel Design Option page.
2. Can I contact 10x support if I have questions about requesting an advanced custom panel design (e.g., feasibility of experimental design, assay sensitivity or specificity for a target gene, multi-species, etc.)?
Submit your questions to support@10xgenomics.com if you have technical questions prior to starting an advanced custom panel design. It is important to carefully consider the feasibility and experimental design before starting this design process.
3. Who do I contact if I have questions about purchasing an advanced custom panel before starting the panel design process?
Contact your local sales representative or distributor for any questions about advanced panel designs. If you do not know your local sales representative, contact customerservice@10xgenomics.com.
4. What is an Advanced Panel Upgrade and how do I pay for it?
An Advanced Panel Upgrade is a product upgrade that enables you to generate panels for applications not possible with standard gene expression panels. The advanced custom and species custom designs are complex and require bespoke design optimization for each panel request. You can upgrade your custom panel to an advanced panel by purchasing an Advanced Panel Upgrade.
You need to have a PO for a custom panel and an Advanced Panel Upgrade associated with the panel design ID in order to submit a request for an advanced custom panel on the Xenium Panel Designer. We will invoice you for both part numbers after the panel design has been finalized (Xenium v1 PNs, Xenium Prime PNs).
5. Why do I need an Advanced Panel Upgrade?
Each advanced custom or species standalone custom request is built by the 10x Genomics Applied Bioinformatics team in collaboration with customers. The Advanced Panel Upgrade is only required for designs led by our Applied Bioinformatics team.
6. I plan to request a human or mouse panel and to add probes for bacterial or viral genes. Should I select "human", "mouse", or "other" for my organism?
We would consider "human" or "mouse" to be the organism in this case, with advanced custom targets added.
7. How do I specify sequences for advanced custom design?
There are a few common target design modes: isoforms, single nucleotide variants, exogenous genes, T cell receptors, and barcode detection. Guidance for submitting information for these targets is provided in the Advanced workflow documentation.