All the same visualizations and tools used to examine T cell receptor (VDJ-T) datasets are also available for B cell. In addition, Loupe V(D)J Browser has a few specific features to accommodate the analysis of B cell immunoglobulin data. For example, Loupe V(D)J Browser can identify IGH, IGK, and IGL immunoglobin chains. Note that the iNKT/MAIT Evidence filter does not apply to B cells.
A PBMC B cell Ig dataset has been provided for you to download and explore. Both V(D)J T and B cell libraries were created from this donor sample, but there is no Antigen Capture library. Cell Ranger v5.0 was used for data analysis.
Immunoglobulin class switch recombination, or isotype switching, occurs when the constant region of a B cell V(D)J heavy chain sequence changes such that the cell produces antibodies of a different isotype. Cells that produce different Ig isotypes may belong to the same clonotype; this is accounted for by the clonotype grouping algorithm in Cell Ranger.
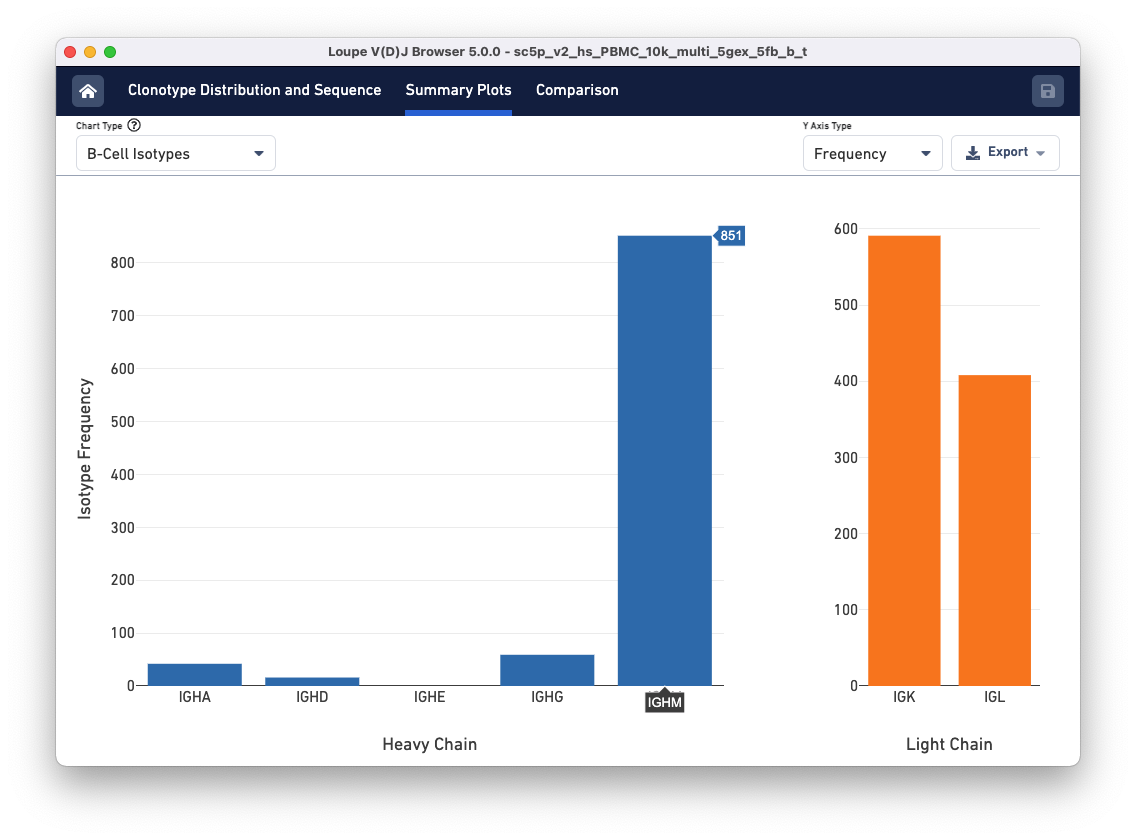
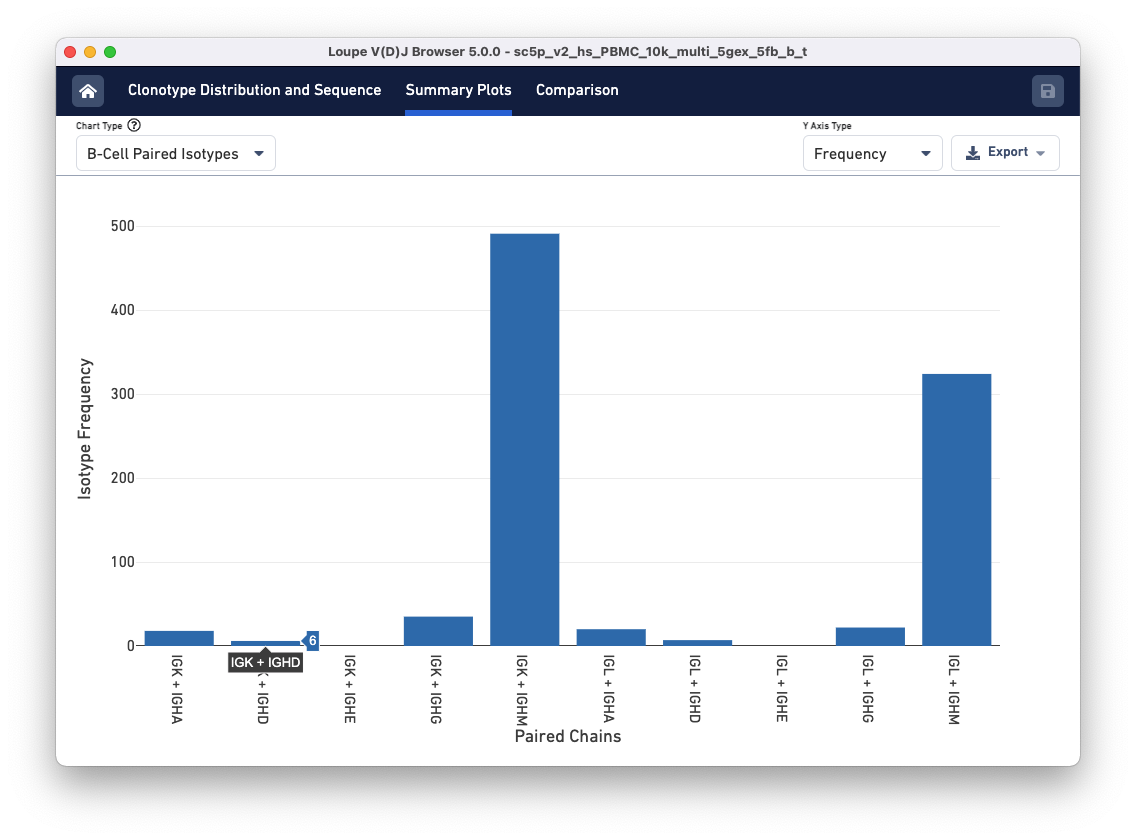
When viewing a B cell sample, two additional summary charts allow you to analyze the isotype distribution. To access these, go to the Summary Plots tab and click the Chart Type drop-down. Scroll to the bottom of the list to find the B Cell Isotypes and B Cell Paired Isotypes.
First, select the B Cell Isotype chart. This chart displays the frequency of immunoglobin isotypes and the type of light chains in the sample.
Next, select the B Cell Paired Isotype chart. The chart displays the distribution of paired isotypes and light chains among clonotypes in the sample. Only clonotypes where there is exactly one heavy chain and one light chain are included in this chart.
Comparing across samples is more powerful, whether those samples are from different time points, different conditions, or different sources. Take a look at the new features that allow Loupe V(D)J Browser to compare multiple samples together.