In this part of the tutorial, we use Loupe V(D)J Browser to look at clonotypes within a gene expression cluster and compare clonotype distributions between clusters. You need these files to proceed:
- Download Loupe V(D)J Browser v5.0
- Download Lung Carcinoma Gene Expression .cloupe file
- Download Lung Carcinoma T Cell .vloupe file
If you are new to Loupe V(D)J Browser, you may want to read the Loupe V(D)J Browser tutorial before continuing.
To start, open Loupe V(D)J Browser, and click on Open Loupe V(D)J File. Choose the LungTumorT_V4 dataset and click Load. The landing page should look like this:
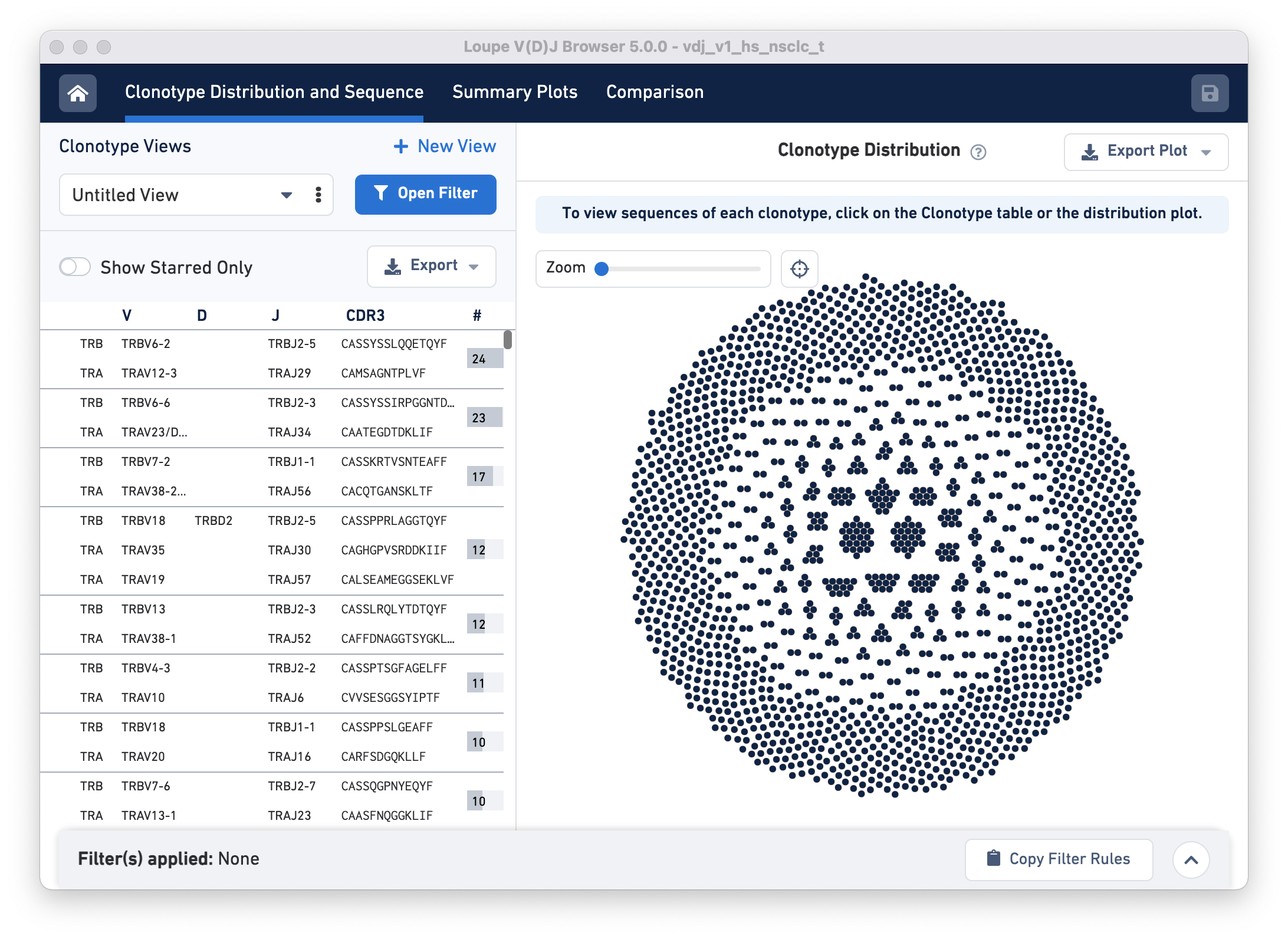
Next, click on the Open Filter button. The pop-up panel has a row called Cluster from an imported Cloupe file. In this row, click on the hyperlink 'Click here to select a cloupe file' and load the LungTumorGEX.cloupe.

The Cluster from an imported Cloupe file row now has a text box drop-down that allows you to filter clonotypes by selecting a cluster that was previously defined in the .cloupe file.
Filter clonotypes that belong to the CD8 cluster by typing 'CD8' in the Enter Cluster Name box. Alternatively, click on the drop-down and scroll until you find the CD8+ Cytotoxic T Cells cluster option in the list. Adding this filter should highlight the CD8+ T cells in the Clonotype Distribution plot.

Clicking on each clonotype in the clonotypes list panel (left) shows you the alignment view for all the exact subclonotypes in that clonotype.
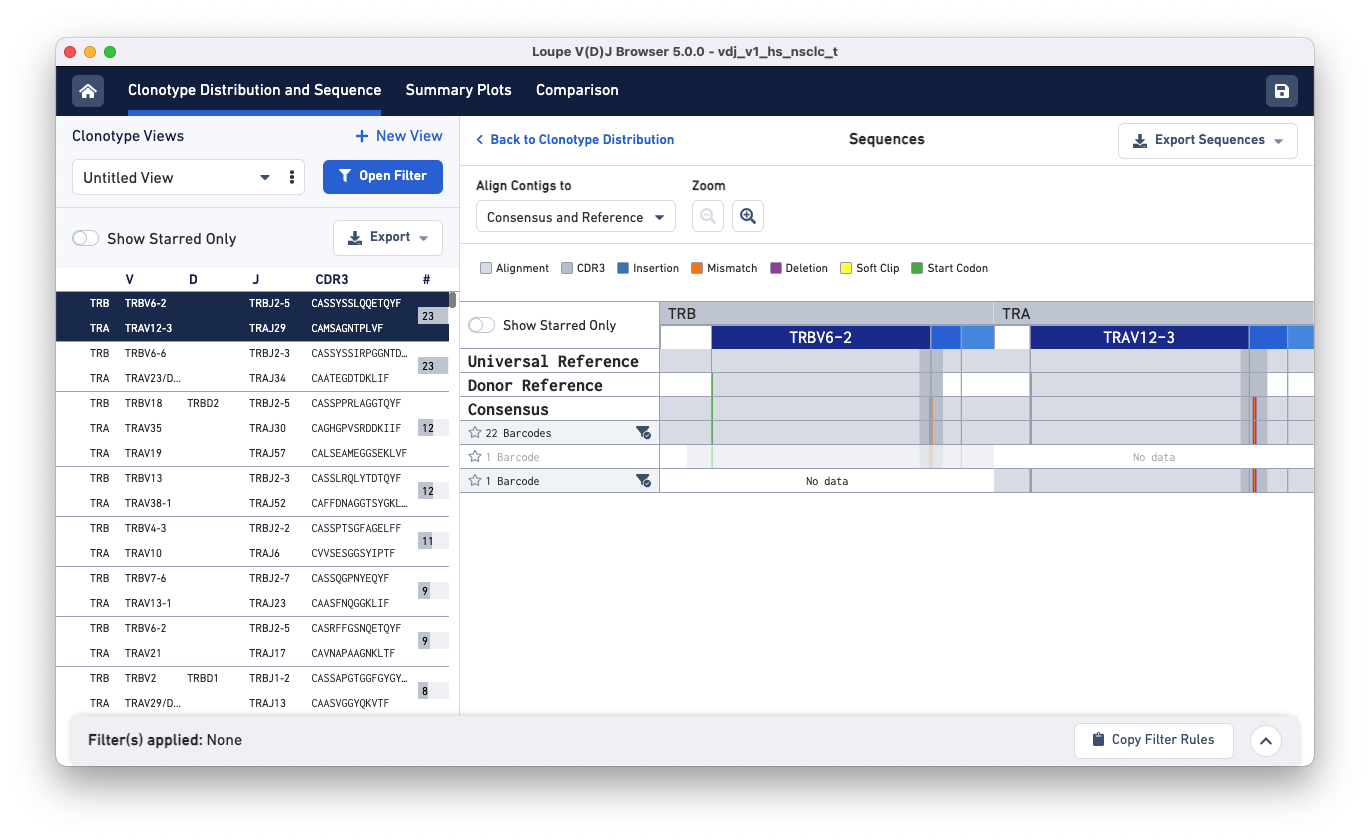
Click on the gray parts of the alignment or the zoom in button to see the nucleotide sequences of the three exact subclonotypes, the universal and donor references, and the consensus.
Now that you have seen how to combine V(D)J and Gene Expression data with Loupe Browser and Loupe V(D)J Browser, you may have ideas about how to use the Chromium™ Single Cell V(D)J Solution for your experiments. If you have any questions or feedback about the software or other parts of the integrated V(D)J and Gene Expression workflow, contact support@10xgenomics.com.