Bug fixes
-
Fixes an issue in aggr where files would fail to be copied on NFSv4 File Systems.
-
Fixes an issue in multi where r1-length and r2-length settings were ignored for
vdj.
Changes that apply to Gene Expression and Feature Barcode analysis
-
Cell Ranger v5.0 introduces a
--no-bamoption that disables the generation of aligned BAMs for gene expression and feature barcode datasets. If you have no need for these files, then disabling their generation can significantly speed up the pipeline. -
Cell Ranger v5.0 introduces upgraded protein aggregation detection and filtering algorithm. By directly using the protein counts, more aggregate GEMs are detected and filtered out before proceeding with cell calling.
-
Cell Ranger v5.0 introduces an
--include-intronsoption for counting intronic reads using 3’ Gene Expression and 5’ Gene Expression products. The usage of pre-mRNA references for counting intronic reads is now deprecated.- The
--include-intronsoption, introduced in Cell Ranger 5.0, works by aligning reads to a normal reference transcriptome with STAR. After alignment, the reads mapping to introns are annotated and counted similarly to reads that are aligned to exons. Previously, the pre-mRNA reference strategy implemented with Cell Ranger 4.0 and earlier involves alignment to a modified reference transcriptome that categorizes intronic regions as exonic. There are slight differences in read alignments produced by the STAR aligner when a pre-mRNA reference is used compared to a normal reference using--include-introns. These differences result in small overall differences in counted UMIs for intron-mode compared to pre-mRNA-reference.
- The
-
Ported a fix from upstream
IRLBAthat fixes incorrect behavior in rare circumstances. -
On some Linux distributions, NFS implementations would surface an improper error during file copy. We have implemented a workaround for our affected native code.
Changes that apply to Gene Expression, Feature Barcode, and V(D)J analysis
-
Cell Ranger 5.0 introduces the multi pipeline that can simultaneously process any combination of 5' Gene Expression, Feature Barcode (cell surface protein or antigen) and V(D)J libraries from a single GEM well. The multi pipeline uses the cell calls provided by the gene expression data to improve the cell calls inferred by the V(D)J library.
-
A new metric, “Number of Short Reads Skipped”, is added to the web summary, indicating the total number of read pairs that were ignored by the pipeline because they do not satisfy the minimum length requirements.
Changes that apply to V(D)J analysis
-
Cell Ranger v5.0 introduces a new clonotype grouping algorithm that computationally approximates groups of cells which are descendants of a single, fully rearranged common ancestor and infers the germline sequence of the V genes from each individual in the dataset. In previous versions (4.0 and earlier), the algorithm grouped cells based only on the set of productive CDR3 nucleotide sequences. As a consequence, whenever a true clonotype had a CDR3 mutation, the true exact subclonotypes were presented by the algorithm as multiple separate clonotypes. The previous approach to clonotyping in Cell Ranger 4.0 and earlier led to inaccuracies in the B cell clonotypes due to the grouping by unique CDR3 sequence. Additionally, single-chain clonotypes were reported as separate clonotypes, which could lead to both over- and under-estimation of the size of a given clonotype. The new clonotyping algorithm is improved in specificity, sensitivity, and overall accuracy because it accounts for mutations found in the V(D)J transcript and in the V(D)J junction. It also merges single chain clonotypes into the correct fully-paired clonotypes for both T cells and B cells. Additional cell filters are also imposed during clonotyping to improve data quality.
-
Changes to V(D)J outputs:
-
The following output files are removed in 5.0:
consensus.fastqandconsensus_annotations.json -
The following output files are added in 5.0: - Contig info binary file, which would be used as an input to aggregate V(D)J samples - Donor reference fasta
-
Two new columns are added to the clonotypes.csv file that displays the iNKT/MAIT evidence.
-
The files
filtered_contig_annotations.csv,filtered_contig.fasta,filtered_contig.fastqnow only contain data from the contigs in cell barcodes that are productive. -
A number of new fields are added to
consensus_annotations.csv:v_start,v_end,v_end_ref,j_start,j_start_ref,j_end,cdr3_start,cdr3_end
-
-
The recommended V(D)J reference packages for human and mouse have been updated from v4.0-5.0. The changes to the V(D)J reference sequences are listed below:
HUMAN:
- Replace IGKV2D-40, whose leader sequence appears to be truncated.
- Delete IGKV2-18, which is probably a pseudogene.
- Delete IGLV5-48, which is truncated on the right.
- Delete TRBV21-1, which has multiple frameshifts.
- Add IGHV4-30-4, which was missing.
- Add IGKV1-NL1, which was missing.
- Add IGHV4-38-2, which was missing.
MOUSE:
- Delete TRAV23, which is frame-shifted.
- Delete the first base of the constant region gene IGHG2B.
- Make a six-base insertion in IGKV12-89, based on empirical data.
- Correct IGHV8-9, whose amino acid sequence showed the canonical C at the end of FWR3 as S. This is consistent with 10x data.
- Add an allele of IGKV2-109, which was missing.
- Add IGKV4-56, which was missing.
- Add IGHV1-2, which was missing.
-
cellranger aggrnow aggregates V(D)J data, allowing users to recompute V(D)J clonotype groupings across the combined data. -
Soft deprecation of
--force-cellsincellranger vdj:-
Since Cell Ranger 3.1, due to filters in the VDJ assembler,
--force-cellsin VDJ pipelines did not behave as users would expect it to behave. Users can only apply--force-cellsto the number of barcodes passing the combined filters in the assembler. -
This makes it effectively impossible for users to increase the number of recovered cells. Rather, it is only possible to reduce the number of recovered cells using
--force-cellsin this context, unlike the behavior in thecellranger countpipeline. -
Because this specific flag is likely to be misunderstood by users, and is also not highly requested, we are starting to deprecate it. In Cell Ranger 5.0,
--force-cellswill be available only as an undocumented silent option. This will also allow users who are using this routinely in their workflows to anticipate eventual deprecation.
-
Changes that apply to Gene Expression and Feature Barcode analysis
-
Targeted Gene Expression analysis is available in Cell Ranger 4.0 and is invoked by specifying the
--target-paneloption when running the cellranger count command. -
Cell Ranger 4.0 introduces the new
targeted-comparepipeline for direct comparative analysis of matched parent Whole Transcriptome Amplification (WTA) and Targeted Gene Expression datasets. -
Cell Ranger 4.0 includes the new
targeted-depthsubcommand to estimate sequencing depths appropriate for Targeted Gene Expression experiments based on input WTA results and an associated target panel file. -
Recommended reference packages for human and mouse have been updated from version 3.0.0 to 2020-A:
-
Transcriptome annotations updated from Ensembl 93 to GENCODE v32 (human) and vM23 (mouse), which are equivalent to Ensembl 98.
-
GRCh38 and mm10 sequences are not changed; chromosome names now follow the GENCODE/UCSC convention (e.g.,
chr1andchrM) rather than the Ensembl convention (1andMT). -
Additional filtering removes genes with unreliable annotations that often overlap more legitimate genes (see build scripts for details), resulting in improved overall sensitivity. 2020-A reference packages are backwards compatible with Cell Ranger v3.1.0 and prior.
-
Mapping rates and gene/UMI sensitivity are increased due to more comprehensive annotations and improved manual curation of genes:
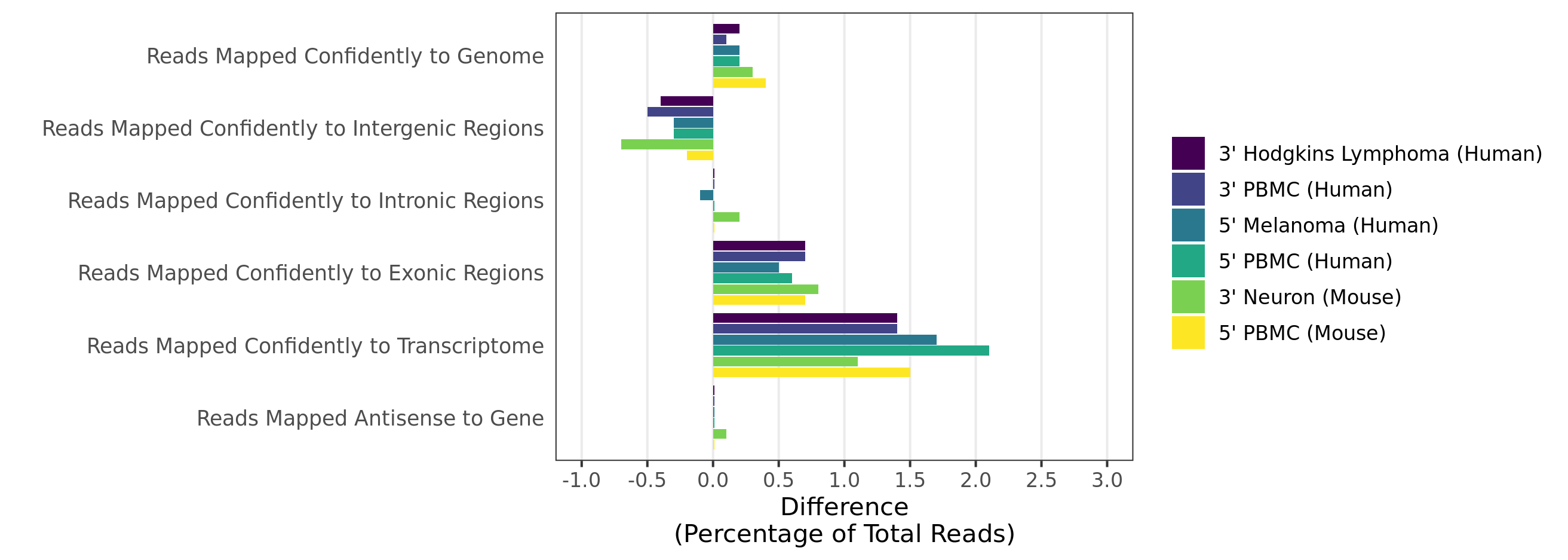
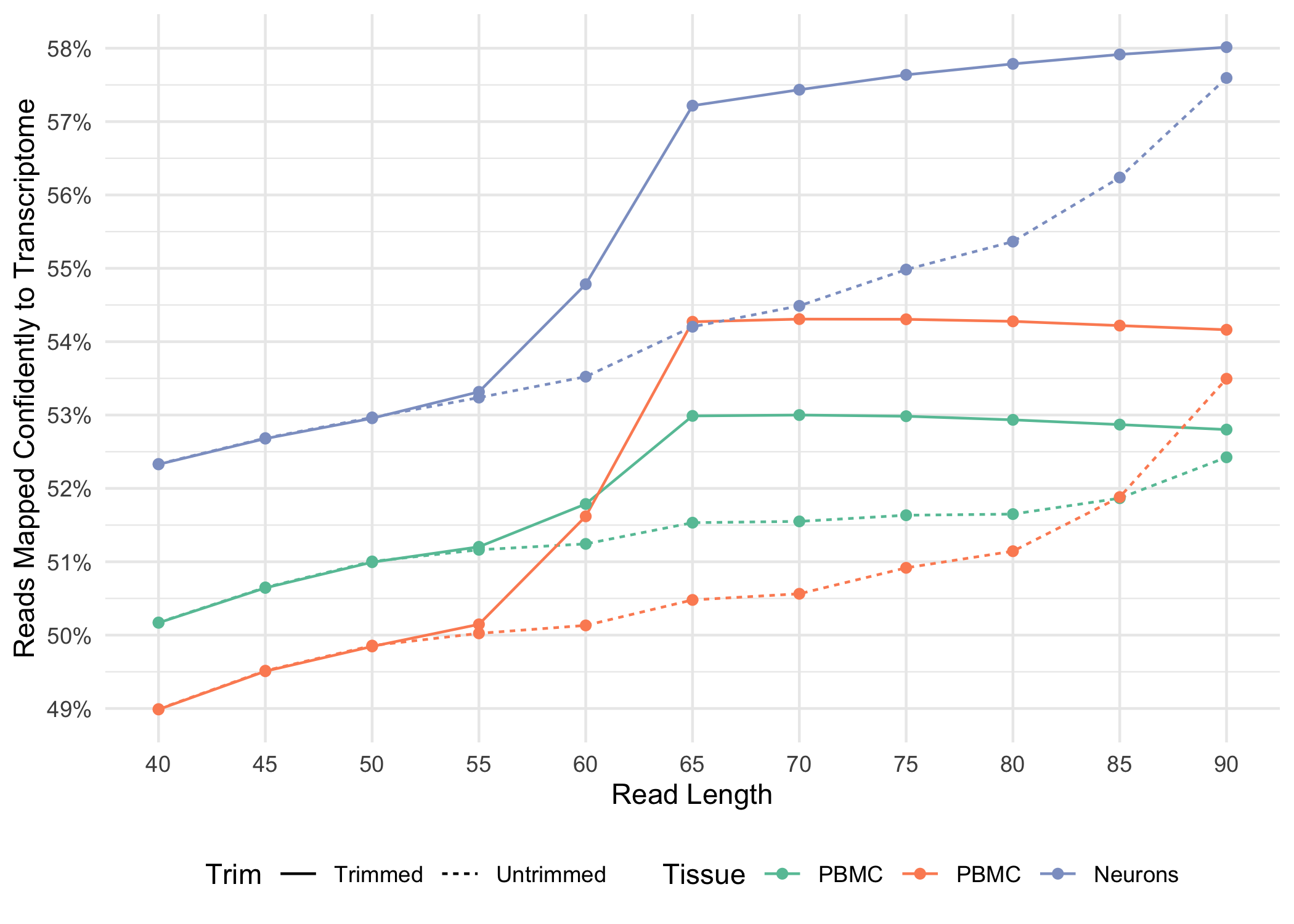
- When analyzing 3’ Gene Expression data, Cell Ranger 4.0 trims the template switch oligo (TSO) sequence from the 5’ end of Read-2 and the poly-A sequence from the 3’ end before aligning reads to the reference transcriptome. This behavior is different from Cell Ranger 3.1, which does not perform any trimming.
A full length cDNA molecule is normally flanked by the 30-bp TSO sequence, AAGCAGTGGTATCAACGCAGAGTACATGGG, at the 5' end and the poly-A sequence at the 3' end. Some fraction of sequencing reads are expected to contain either or both of these sequences, depending on the fragment size distribution of the library. Reads derived from short RNA molecules are more likely to contain either or both TSO and poly-A sequence than longer RNA molecules.
Trimming results in better alignment, with the fraction of reads mapped to a gene increasing by up to 1.5%, because the presence of non-template sequence in the form of either TSO or poly-A confounds read mapping. Trimming improves the sensitivity of the assay as well as the computational efficiency of the pipeline. Tags ts:i and pa:i in the output BAM files indicate the number of TSO nucleotides trimmed from the 5' end of Read-2 and the number of poly-A nucleotides trimmed from the 3' end. The trimmed bases are present in the sequence of the BAM record and are soft clipped in the CIGAR string.
Below, we illustrate how the fraction of reads mapped confidently to the transcriptome varies for both trimmed and untrimmed alignment as a function of read-length for a variety of sample types .
-
Cell Ranger 4.0 adds support for an “un-tethered” Feature Barcode pattern, (BC) without an anchor, specified in the Feature Reference CSV. This option allows the user to specify the sequence of the Feature Barcode without specifying a particular location on the read where the sequence is expected to be found.
-
cellranger reanalyzenow outputs the count matrix used in the analysis, so as to reflect any subsetting of barcodes used. -
Bug fixes for GTF files output by
mkref. These changes do not affect the pipeline results.- GTF attributes with duplicate keys (e.g., tag
"value1";tag "value2";) are handled correctly. Previously, only the last such attribute was kept. - GTF attributes with unquoted integer values (e.g.,
exon_number 1;) are kept. Previously, they were removed. - GTF lines end with semicolons.
- Unix line endings are used rather than DOS line endings, consistent with other Cell Ranger outputs.
- GTF attributes with duplicate keys (e.g., tag
-
Bug fixes for the BAM file
- The duplicate flag (
0x400) is set correctly in the secondary alignments (flag0x100) of PCR duplicate reads and low-support UMI reads (xf:i:2) - Low-support UMI reads (
xf:i:2) have the corrected barcode in UB:Z. Previously, it contained the raw barcode.
- The duplicate flag (
-
BAM file changes
- Cell Ranger v4.0 will not output the
li:itag. TheRG:Ztag contains this information. - Cell Ranger v4.0 will not output the
BC:ZandQT:Ztags.
- Cell Ranger v4.0 will not output the
-
Cell Ranger v4.0 now relies on Orbit to perform transcriptome alignment, which leverages a modified STAR v2.7.2a. These modifications provide compatibility with “versionGenome 20201” references, such as those generated by STAR v2.5.1b. In Cell Ranger 4.0 we still provide and use STAR v2.5.1b for other purposes such as
cellranger mkref. In our testing we did not note any differences in transcriptome alignments between the STAR shipped in Cell Ranger 3.1 (STAR v2.5.1b), STAR v2.7.2a, or Orbit. -
mkfastqnow accepts file names without lane number, e.g.,sample1_S1_R1_001.fastq.gz. -
Cell Ranger's
aggrpipeline no longer supports the aggregation of v1mol_info.h5files.
Changes that apply to Gene Expression, Feature Barcode, and V(D)J analysis
-
mkfastqsupports dual-indexed libraries for gene expression, both WTA and Targeted, V(D)J, and Feature Barcode datasets. -
mkfastqsupports a new sequencing configuration for Novaseq where the I2 index may need to be reverse-complemented before demultiplexing dual-indexed libraries. -
mkfastqnow accepts file names without lane number, e.g., sample1_S1_R1_001.fastq.gz. -
countandvdjrun approximately two to four times faster than in Cell Ranger v3.1, depending on the sequencing data, and reduces disk I/O by half. -
A new command-line interface with improved error-handling has been engineered into Cell Ranger v4.0.
-
The Martian pipeline framework has been upgraded to v4.0.
mrpandmrjobwill shut down if they detect that their log files were deleted or renamed. See the Martian release notes for more details. -
The following features present in Cell Ranger v3.1 are no longer present in Cell Ranger 4.0:
mkfastqno longer supports data from the Single Cell 3′ v1 chemistry.- The
cellranger demuxsubcommand has been removed. - The command-line interface does not accept FASTQs created by the deprecated cellranger demux pipeline. If you need to process FASTQs in this layout, contact support@10xgenomics.com for assistance.
cellranger countandcellranger vdjare no longer able to process data from multiple gem-wells through manual editing of MRO files. The Single Cell 3′ v1 and Single Cell 5′-R1 assay configurations will no longer be autodetected in Cell Ranger 4.0. Users who want to analyze data from those chemistries must explicitly specify the chemistry (SC3Pv1orSC5P-R1respectively) using the--chemistryargument.- The
--idargument used by the pipelines has a 64 character limit in Cell Ranger 4.0.
-
The
--idargument used by the pipelines has a 64 character limit in Cell Ranger 4.0.
Changes that apply to V(D)J analysis
-
Recommended VDJ reference packages for human and mouse have been updated from version 3.1.0 to 4.0.0. The changes to the VDJ reference sequences are listed below:
- Remove the first base of the C region in certain cases. In these cases we observe that in most transcripts, the J region and C region overlap by exactly one base.
- Add an allele of the gene IGHJ6 to the human VDJ reference.
-
Bug fix in contig annotation: If a reference D region matches a contig perfectly, annotate the contig with that D region.
-
The command line argument
--chainis added back in 4.0 for rare cases when the automatic chain detection fails. -
A new output
airr_rearrangement.tsvis added, which contains annotated contigs of VDJ rearrangements in the AIRR TSV format. -
The VDJ reference is copied to the outputs folder starting with Cell Ranger v4.0.
-
Feature Barcoding Only Analysis - It is now possible to run
cellranger countusing Cell Surface Protein (antibody captured) libraries without a GEX library. The previous version of Cell Ranger required a Gene Expression library along with a library generated by Feature Barcoding technology. However, the new version of Cell Ranger provides customers with flexibility to sequence either one of the libraries, or both. In particular, cell calling now works with antibody counts only, and all secondary analyses (PCA, t-SNE, UMAP, clusterings) work with antibody-only count matrix as well. -
UMAP based lower dimensionality projections of datasets analyzed by
cellranger countare now produced in addition to the previously produced t-SNE projections. The projections are made available both as CSV files and as data that can be directly viewed in Loupe Browser. The parameters for the projection can also be modified and experimented with using cellranger reanalyze. This alternate visualization method has become increasingly popular for visualizing single cell data since the earliest report that used it. For more details, see the description in the algorithms overview section. -
New Web Summary Look - The Cell Ranger
web_summary.htmlfile has been updated to match the styles and formats of other 10x products. Compared to the old version users will notice new fonts and some aesthetic changes in the new version. -
Bug Fix: If equal numbers of reads with given Barcode / UMI combination map to two genes, the assignment of the Barcode / UMI are now considered ambiguous and not reported in moleculeinfo.h5 or the count matrix. Previously they were reported _twice, once for each gene.
-
Other minor bug fixes
Release Notes for Martian 3.2.3: Job Scheduling
-
Fix a crash in cases where the
mrpbinary becomes unavailable on disk during a pipestance run. -
In addition to logging the type of filesystem for the pipestance directory, mrp will also log the type of filesystem for the martian bin directory (which is often different from the pipestance directory), and also the mount options for both directories.
-
Regardless of
--jobintervalsetting,mrpwill now never attempt to submit more than one job at a time to the queue in cluster mode. -
mrpwill now shut down if the pipestance log file has been deleted, even if a new one has been created in its place. This prevents problems in the case where the pipestance directory (including the log and lock files) have been deleted. -
Memory cgroups limits are now detected, reported, and used as default limits where applicable. This should be especially helpful for users submitting
mrpto a cluster such as SLURM which uses memory cgroups to prevent jobs from using too much memory, by preventingmrpfrom trying to use more than the job's allowance. -
Other small bug fixes and performance improvements.
V(D)J Release Notes
Major algorithm changes and effects on performance
-
The assembly, annotation and cell calling algorithms have all been replaced, as have the reference sequences. However with noted exceptions, the interface is unchanged.
-
Many changes were made to the assembly algorithm that allow it to achieve the same sensitivity using less data. After these changes, the recommended sequencing configuration was changed to 26 x 91 (from 150 x 150), while leaving the number of read pairs per cell fixed at 5000. This enables V(D)J, Gene Expression and Feature Barcoding libraries to be sequenced in a single run, thereby simplifying the workflow.
-
The effect of the new changes varies considerably from sample to sample and we have added a discussion on Experimental Design that explains some of this. In some instances the number of productive pairs increases markedly if the same dataset is rerun with the new code.
-
The old read configuration 150 x 150 is still supported and may be preferable for some users, because of pricing or availability, particularly for users who are running only V(D)J data. For 150 x 150, the recommended depth is proportionally lower, 2000 read pairs per cell.
-
Many corrections were made to the Prebuilt reference sequences.
-
Contig annotation has been improved in several ways. This includes more accurate detection of CDR3 regions, a more stringent full-length requirement, and a requirement that V segments begin with a start codon (coupled to reference sequence corrections). This could affect annotation for species other than human or mouse, having incomplete reference sequences.
-
A productive pair is no longer declared in cases where there are three or more contigs having the same chain type (e.g. TRB, TRB, TRB). In such cases the GEM may contain two or more cells.
-
Some new large clones are now reported, that were missed previously for a variety of reasons, including failure to align J segments having high somatic hypermutation.
-
A productive pair is no longer declared in cases where three or more contigs share the same chain type (e.g. TRB, TRB, TRB). In such cases the GEM may contain two or more cells. In addition, certain clonal expansions of plasma cells are now contracted because the expansion represents mRNA leakage during processing, rather than a true biological expansion. Finally, requirements for small clones sharing a chain with a large clone have been tightened to reduce the likelihood of false clones arising from ambient mRNA or doublets. All of these changes correctly reduce the number of reported productive pairs (usually by a small fraction).
-
Because of these changes, we recommend that customers rerun existing datasets using Cell Ranger 3.1 if possible.
-
Because cell calling is changed, the denominator used for computing the Cells With Productive V-J Spanning Pair metric may have changed. For this reason, differences in performance between Cell Ranger 3.0 and 3.1 are better assessed using the Number of Cells With Productive V-J Spanning Pair metric.
-
Cell Ranger 3.1 is significantly faster. There are five fewer stages in the pipeline.
Interface Changes:
-
Cell Count Confidence is no longer reported because we found that in some cases incorrect counts were reported with high confidence. Cell counting from V(D)J data alone is limited in accuracy because targeted cells having sufficiently low expression cannot be detected.
-
Contigs Unannotated is no longer reported because all contigs are now annotated. The justification for this is that since enrichment uses primers binding to constant regions, bona fide contigs would be expected to have at least a C annotation.
-
For species other than human or mouse, for which custom primers are needed, the sequences of the inner enrichment primers must now be supplied as a command-line argument.
Job Scheduling Changes
- Add support for SGE and LSF clusters that track virtual memory use.
Enable Analysis of CITE-seq Experiments
-
Cell Ranger can now process data from experiments where the antibodies were conjugated to oligonucleotides that were captured by oligo-dT primers. Previously, only experiments which used the Chromium Single Cell 3' Feature Barcode Library Kit, which utilizes a different capture sequence for Gene Expression and Feature Barcoding data, could be analyzed.
-
Please note that while Cell Ranger is now compatible with CITE-seq data, CITE-seq is not a supported application. To ensure full support for your 10x data analysis please visit the Feature Barcode Analysis page to see the supported Feature Barcoding technology.
Bug fixes
-
Fix an issue where STAR would crash on CPUs without AVX support.
-
Fix a determinism issue when aggregating 3' v2 and v3 data.
-
Increase the memory reservation for the SORT_BY_POS stage.
General
- Cell Ranger has been overhauled to support user-defined Feature Barcoding reagents, and to quantify these features alongside standard gene-expression reads. See Feature Barcoding for details. For users who have already run their data through earlier versions, there is no need to rerun it again using this new version.
Cell Calling Changes
-
Cell Ranger 3.0 implements a version of the EmptyDrops cell calling algorithm that will call more low RNA content cells, especially when they are mixed with a population of high RNA content cells. See Cell Calling Algorithms for details.
-
The cell calling 'knee-plot' in the web summary now indicates what fraction of barcodes in each segment of the curve were called as cells, since the new cell calling algorithm no longer makes a hard threshold on UMI counts.
Output File Format Changes
-
The file formats of the gene-barcode matrix (now called the feature-barcode matrix) have changed to accommodate Feature Barcoding results.
-
The mtx and barcodes.tsv files are now gzipped to save disk space The genes.tsv file has been renamed features.tsv.gz, and contains extra columns indicating the
feature_typeof each gene / feature. -
See Feature-Barcode Matrices for details.
-
As part of this change,
cellranger-rkitis deprecated. We recommend Seurat for analysis in R. -
The Molecule info file format has been substantially changed to enable output from the new Feature Barcoding technology and remove rarely used mapping metrics.
Cell Ranger 2.2.0 will require CentOS/RedHat 6 or Ubuntu 12 or later. See the 10x OS Support page for further information.
-
Fix Martian UI display in FireFox
-
Fix non-integral resource requests (memory/threads)
-
Fix SUBSAMPLE_READS producing wrong metric names. Newer version of Martian no longer casts zero-fractional floats to ints, which this code was relying on to produce metric names with integral subsampling rates in them.
-
Fix failure to detect inclusion list with demux when a single Sample Index is bad
-
Fix always-on multi-chromosome transcript warning in
mkref -
Fix stall in ALIGN_READS on filesystems that don't support named pipes
-
Fix python error when autodetect of chemistry fails with multiple FASTQ paths
-
Fix handling of sample names with multiple underscores in
mkfastqpipeline -
Fix suppression of process limit errors in the
mkfastqQC stage
Changes to mkfastq
-
Barcode-aware QC stage is now opt-in via the
--qcflag. -
Limit total CPU usage across stages to 12 cores unless
--localcoresis specified. This should improve reliability on machines with high numbers of cores.
Cell Ranger 2.1.1 Gene Expression
Note: This is expected to be the last version of Cell Ranger to support CentOS/RedHat 5 and Ubuntu 10. If you are using one of those operating systems, Cell Ranger will now warn you. Future versions of Cell Ranger will require CentOS/RedHat 6 or Ubuntu 12 or later. See the 10x OS Support page for further information.
Bug Fixes
-
Fix library ID labels being out of order in the matrix HDF5 file produced by
cellranger aggrwhen 10 or more libraries are aggregated. This manifests as Loupe Cell Browser showing the library ID labels out of order after runningcellranger reanalyze. -
Fix an
out-of-memoryerror occurring when generating the kmer index on a reference with very long transcripts, e.g. on a pre-mRNA reference used when analyzing nuclei samples. -
Fix crash when analyzing FASTQs produced by SRA's
fastq-dump. -
Fix the Differential Expression table in the web summary disappearing when gene IDs are equal to gene names in the reference GTF.
-
Fix a few web summary metrics becoming negative when more than 2.1 billion reads are analyzed at once.
-
Fix incorrect parsing of the
--localcoresargument, causing--localmemto be ignored when specified immediately after--localcores. -
Fix crash in
mkfastqon NovaSeq when RunParameters.xml is namedrunParameters.xml. -
Fix hang when running
sitecheckon some systems. -
Fix error reporting in python stage code imports.
-
Fix estimation of stage virtual memory usage.
Improvements
-
Truncate large metadata files when generating a tarball for upload to 10x, rather than omitting them. Remove the requirement that the reference FASTA file modification time precede the STAR index file modification times.
-
The default
--localmemin cluster mode will no longer ever be more than the free memory available when thecellrangerstarts.
New Features
-
Add support for and autodetection of Single Cell 5' gene expression libraries, with support for both paired-end alignment (150x150) and R2-only alignment (26x98).
-
Add
--r1-lengthand--r2-lengthoptions tocellranger countwhich enable hard trimming of input FASTQs. -
Add
--exclude-genesoption to cellranger reanalyze which, analogously to--genes, allows for the exclusion of some genes from the secondary analysis (PCA, clustering, etc.). -
Add
--chemistrytocellranger countto override the automatic chemistry detection.
Performance Improvements
-
Reduce the run time by 30%.
-
Reduce the disk storage high-water-mark by 60%.
Algorithm Improvements
- Change the Antisense Reads Metric to only count a read as antisense if it has no sense alignments, effectively prioritizing sense alignments over antisense for this computation.
Output File Changes
-
Stop generating the TR and TQ BAM tags because these tags were retaining trimmed sequences that Cell Ranger would ignore anyway after converting the BAM back to FASTQ.
-
Add more mapping metrics (Reads Mapped to Genome, Reads Mapped Confidently to Genome), and reorder the mapping metrics to be consistent with their order of computation.
Bug Fixes
-
Fix mkfastq allowing max
bcl2fastqthreads to exceed--localcores. -
Fix mkfastq crashing when reading NovaSeq quality data from RTA 3.3 and later.
-
Fix excessive memory requests in
SC_RNA_ANALYZER. -
Fix nondetection of louvain binary failure in
RUN_GRAPH_CLUSTERING. -
Fix crash in
RUN_GRAPH_CLUSTERINGwhen/dev/stdindoesn't exist. -
Fix the barcode rank plot concatenating instead of unioning barcodes in multi-genome datasets.
System Requirements Changes
- Cell Ranger no longer supports Ubuntu 8 or CentOS 5.2 Linux distributions. Ubuntu 10.04 LTS or CentOS 5.5 or greater are now required.
Job Scheduling
-
The pipeline management system, mrp, is now open source on GitHub.
-
The monitoring port for the user interface is now always on by default, with an OS-selected port if none is specified.
- This behavior can be disabled with
--disable-ui. - Access to the user interface port, if no port was specified explicitly, now requires a randomly-generated authentication token. This token is visible in the pipeline standard output and in the
_uiportfile.
- This behavior can be disabled with
-
A new tool,
mrstatis now available.- Given the path to the directory with a running pipeline, mrstat will return basic information about the progress of the pipeline.
- With the
--stopflag, it will cause the pipeline to fail and exit.
-
Two new variables are available for use in cluster-mode templates:
__MRO_JOB_WORKDIR__can be used to specify the absolute path to the directory where the job should execute. This should alleviate issues on clusters such as PBS which sometimes do not set the working directory correctly.__MRO_ACCOUNT__passes theMRO_ACCOUNTenvironment variable frommrp's environment. This is intended for cluster managers which support charging resources to specific accounts.
-
The pipeline standard output and log will now periodically provide progress updates for in-progress stages.
-
mrpwill now provide more clear and useful error reporting when the pipeline directory runs out of disk space. -
Several enhancements to the reliability of pipeline restart.
-
Fixes for several cases where a pipeline could "hang" indefinitely without making further progress.
-
Pipelines should now do a better job of staying within their CPU usage allocation.
Bug fixes
- Properly ignore SIGHUP when a pipeline is run using nohup.
Pipeline Argument Changes
-
Add
--overrideoption to all pipelines, allowing for stage-level overrides for cores and memory. -
Reanalyze no longer requires
--aggto persist library ID; it is only required for persisting user-defined fields.
Bug fixes
-
Fix CHUNK_READS using more cores and using them less efficiently than intended.
-
Fix
aggrusing incorrect downsampling rates when more than 10 libraries are aggregated. -
Fix mkfastq proceeding even after
bcl2fastqis killed. -
Fix lack of robustness to rare events where NFS latency induces double file deletion or double directory creation events.
-
Fix ALIGN_READS proceeding after the STAR subprocess fails, causing crashes in
ATTACH_BCS_AND_UMIS. -
Improve error messages when STAR or samtools fail in ALIGN_READS.
-
Fix spaces in transcript IDs causing
ATTACH_BCS_AND_UMISto crash. mkref no longer allows spaces in transcript IDs. -
Fix crash when reads are adapter-trimmed by
bcl2fastqand some reads end up empty. -
Fix out-of-memory condition in
ATTACH_BCS_AND_UMISfor some libraries with >800M reads. -
Fix question marks replacing axis titles of barcode rank plot in web summary.
-
Fix excessive memory consumption and runtime of
mkfastqon large sample sheets.
Job Scheduling
-
Fix several cases where, after
mrp(which is invoked bycellranger) gets killed, it was not able to restart correctly. -
On SGE clusters,
cellranger/mrpnow periodically runs qstat to verify that the jobs it queued have not been killed or canceled. -
If the run fails, instead of just displaying a message pointing the user to the relevant
_errorsfile, the contents of that file is printed.
-On automatic retry of failed stages, the reason for the original failure is logged.
mrp is now more resilient against certain kinds of filesystem errors.
-
In the event of certain types of filesystem problems (such as permissions errors or disk quota),
mrp/cellranger should now sometimes be able to provide more useful and immediate error messages. -
Additional information about the environment cellranger runs in is now logged and included in
mri.tgz. -
Additional information about the environment the analysis runs in is now logged and included in
mri.tgz. -
mrpnow correctly handles the signals sent by SGE and LSF when a soft time limit is reached (e.g. for SGE,-l s_rt 23:00:00). -
Now supports
--overridesmethod to dynamically change additional CPU and memory per stage.
Pipeline argument changes
-
Add
--barcodesand--genesoptions to reanalyze, which allow selection of a specific subset of barcodes and/or genes to use in the secondary analysis. -
Add
--force-cellsoption tocountandreanalyzeto explicitly set the cell count. If specified, Cell Ranger will take the top N barcodes (by UMI count) as cells instead of doing dynamic cell count estimation. -
Rename the estimated cells option from
--cellsto--expect-cellsfor clarity. -
Add
--nosecondaryflag to count, which skips the secondary analysis. Disallow slashes in the--genomeargument inmkref.
Add --id option to mkfastq which allows you to name the output directory.
New subcommands
- Add
cellranger mat2csvcommand, which converts a Cell Ranger sparse gene-barcode matrix to a dense CSV format. Note that the resulting file will be very large, even for a few hundred cells.
Web summary changes
-
Add "Reads Mapped Antisense to Gene" metric, which quantifies reads that are mapped to the non-coding strand of a gene. High values can indicate the use of an unsupported chemistry type, e.g. passing a Single Cell V(D)J library to
cellranger count. -
Add "Fraction GEMs with >1 Cell (Lower / Upper Bound)" metrics, which define a confidence interval for the multiplet rate estimate in multi-genome samples.
-
Add more details to various metric descriptions.
Algorithm improvements
-
Add the requirement that reads overlap annotated exons by at least 50% in order to be considered exonic. As a result, "Reads Mapped Confidently to Exonic Regions" may differ slightly from previous versions.
-
Reduce
EXTRACT_READSper-read runtime by 50% by avoiding OrderedDict and caching metric calculations. -
Reduce
SUBSAMPLE_READSruntime by reducing the number of fixed target values for subsampling (to just 25k and 50k reads per cell).
File format improvements
-
Due to a format change (removal of the IntervalTree object), references produced with
cellranger mkrefusing Cell Ranger v2.0 are not compatible with pipelines from Cell Ranger v1.x. -
Modify the
TX,GX, andGNtags to have more granular transcript/gene annotations. Each BAM record is only annotated with transcripts/genes specific to that alignment, instead of combining annotations from all alignments of the corresponding read. -
Add
REtag, which indicates whether an alignment is exonic, intronic or intergenic.
Bug fixes
-
Fix rare bug in interval arithmetic, leading to exonic reads being falsely annotated as intronic or intergenic. As a result of this bugfix, "Reads Mapped Confidently to Exonic Regions" may differ slightly from previous versions.
-
Fix excessive
EXTRACT_READSruntime (10+ hours) on very large FASTQs such as those produced by mkfastq. -
Fix a crash in
RUN_GRAPH_CLUSTERINGon filesystems that do not support named pipes. -
Fix
SUBSAMPLE_READSusing more VMEM than expected, causing it to be killed by SGE when exceeding the h_vmem limit on certain clusters. -
Fix
mkfastqnot merging output files properly due to sample numbering issues. -
Fix
mkfastqcrash due to-d(demultiplexing-threads) argument being deprecated inbcl2fastq2.19. -
Fix the components.csv file produced by PCA, which did not contain the correct matrix.
-
Fix a crash in RUN_PCA when the number of nonzero genes is smaller than the number of principal components.
-
Fix a crash in mkref with very large genomes; use the
limitGenomeGenerateRAMoption in STAR to overcome its default reference size limit. -
Fix certain special characters (like dashes) in reference names breaking the subsampled genes detected plot.
-
Fix
mkloupedisplaying an unhelpful error message when run on mixed-species runs and those from Cell Ranger v1.1 or earlier. -
Fix the
open-file-handle-limitcheck using the submit host rather than the execution machine. -
Fix
cellranger aggrallowing duplicatelibrary_ids. -
Fix
CLOUPE_PREPROCESStaking the full matrix even afterreanalyzesubselects barcodes. -
Fix a crash in mkfastq on
RunInfo.xmlfiles produced by the NovaSeq. -
Fix a crash in mkfastq when
bcl2fastq2.19 is used in cluster mode or with the--demultiplexing-threadsargument. -
Fix
mkfastqsometimes not properly merging samples inbcl2fastq2.18 and 2.19 due to a change in the order in which lanes are processed bybcl2fastq.
Martian Runtime Changes
- Add caching for deserialized JSON metadata. This improves performance for stages with many chunks.
Miscellaneous
-
Update samtools from 0.1.19 to 1.4.
-
Rename
RUN_PREPROCESStoPREPROCESS_MATRIXin theSC_RNA_ANALYZERpipeline. -
Add
alerts.jsonas an output of theSUMMARIZE_REPORTSstage. This file is a machine-readable list of any abnormal metric values that raised alarms in the web summary. -
For multi-genome samples, display the full reference name rather than a comma delimited list of genomes in the web summary ("hg19, mm10" becomes "hg19_and_mm10").
- Fixes issue preventing mkfastq from demultiplexing data from recent sequencer software versions.
Analysis Improvements
-
Confidently align more reads to the transcriptome, greatly improving alignment rates with shorter reads. - Reads Confidently Mapped to Transcriptome increases from 55% to 62% with 98bp reads and from 34% to 54% with 32bp reads (Human PBMCs vs GRCh38).
-
Add a graph-based clustering algorithm: Louvain Modularity Optimization, which, unlike K-Means, does not require pre-specifying K.
Visualization
-
Automatically produce Loupe Cell Browser (.cloupe) files in the
count,aggregate, andreanalyzepipelines. -
Output a web summary HTML file in the
reanalyzepipeline. -
Be explicit about pre- and post- depth normalization metric values in the
aggrweb summary. -
When the web summary subselects 10e3 cells for display, show the original cluster sizes and not the subselected sizes.
-
Make the web summary HTML slightly smaller by rounding t-SNE coordinates.
-
Update plotly to enable scrollable legends.
File format improvements
- Add Read Group (RG) headers and tags to the output BAM file for better data provenance.
Bug fixes
-
Preserve trimmed bases via the TR/TQ BAM tags for much longer read lengths without crashing.
-
Fix crash when copying files on certain types of network shares that do not support file permissions.
-
Omit no-call bases from Q30 metrics to be consistent with Illumina's Q30 calculation.
-
Allow generation of 3-d (alongside 2-d) t-SNE projections without crashing.
-
Do a better job of hiding dynamic elements while the web summary HTML is loading.
General
-
Make the
--paramsargument to reanalyze optional to enable re-runs with the default parameters. -
Check for mismatches between the library IDs given in the
aggrCSV and those in the matrix file. -
Limit
max_clustersfor K-Means to 50 to ensure sane memory consumption.
-
Fix incorrect results being produced when
aggrprocesses acountoutput that contains multiple libraries (gem groups). -
Exclude untested genes from p-value adjustment.
-
Don't crash when extra commas are present in an IEM samplesheet for
mkfastq. -
Don't crash when no project folders are present for
mkfastq. -
Correctly handle the second index when
mkfastqreceives a dual-indexed IEM samplesheet. -
Allow matrices to have more than 2^31-1 nonzero entries in the matrix HDF5 format.
-
Don't display alerts until the web summary page fully loads.
General
-
Rename main pipeline to cellranger count, which produces a gene-barcode matrix for one library sequenced one or more times.
-
Add support for and autodetection of Chromium Single Cell 3' v2 chemistry; still compatible with v1 chemistry.
-
Fix incorrect default cell count being used when "expected recovered cells" not specified.
New aggr aggregation pipeline
-
New pipeline
cellranger aggrwhich aggregates data from multiple libraries into one dataset. -
Supports combining libraries totalling up to 1,000,000 cells and secondary analysis of the combined data.
-
Automatically performs sequencing depth-normalization for all combined libraries.
New reanalyze custom reanalysis pipeline
- Reruns secondary analysis (dimensionality reduction, clustering, and differential expression) with fully customizable parameters.
New mkfastq demultiplexing pipeline
-
Easier to integrate with existing
bcl2fastq-based workflows. -
Now the preferred demultiplexing method; demux still available but deprecated.
-
mkfastqis a thin wrapper aroundbcl2fastqwith same basic interface. -
Accepts Illumina Experiment Manager-compatible sample sheets with support for 10x sample index sets.
-
Produces FASTQ files and folders in the same structure as
bcl2fastq. -
Generates InterOp output for SAV.
-
Also generates 10x-specific run QC metrics in JSON format.
Scalability enhancements
-
Support combined secondary analysis (dimensionality reduction, clustering, differential expression, and visualization) of up to 1,000,000 cells in under 12 hours with 64 GB of RAM.
-
Change PCA implementation to the Netflix-scale memory-efficient method IRLBA.
-
Decrease runtime of t-SNE implementation.
Analysis Improvements
-
Change differential expression algorithm to the negative-binomial based method sSeq.
-
Report log2 fold-change and p-value for all genes in all clusters.
Sample and genome support
- Add pre-built GRCh38 reference package
Web summary enhancements
-
Add plots that show Sequencing Saturation and Median Genes Detected as a function of downsampled reads per cell.
-
Add Total Genes Detected.
-
Rename "cDNA PCR Duplication" to "Sequencing Saturation."
-
Add chemistry field.
-
Order clusters by size.
-
Add help bubbles to charts.
File format improvements
-
Generate BAM index files with the same basename as the main file.
-
Change cell-barcode and UMI quality tags to CY and UY for better compatibility with the SAM specification.
-
Add TR, TQ tags to BAM to enable lossless BAM to FASTQ conversion.
-
Output HDF5-based sparse matrices in addition to the Matrix Exchange format files for better scalability to high cell counts.
-
Report proportion of variance explained for each principal component.
Martian runtime
-
Pipestance output files (outs) are no longer symlinks.
-
Partial stage restart.
-
Add output filename override, supports two output files having same basename.
-
Add
--onfinishhandler support. -
Add support for units of KB and B for memory reservation in cluster job templates.
-
Pipestances now generate a UUID in _uuid.
-
Add auto-retry mechanism when pipeline stages fail due to causes that appear to be transient.
-
--maxjobs now defaults to 64 in local jobmode. -
--jobintervalnow defaults to 100ms in local jobmode. -
Fix for rare race condition in some Python components
-
Enabled STAR multithreading
-
Added more detailed reference metadata
-
Fixed chromosome name mismatches in 10x reference data
-
Fixed t-SNE algorithm not converging for samples with high cell counts
-
Fixed cell-barcode correction not correcting as many sequences as it should
-
Fixed out-of-memory crash in COUNT_GENES for high-depth samples
-
Fixed occasional loss of the last few reads per chunk in ATTACH_BCS_AND_UMI
-
Added "Reads Mapped Confidently to Exonic Regions" metric to the summary.
-
Changed alert for "Reads Mapped Confidently to Transcriptome" to reflect shorter read lengths and non-human references.
-
Fixed problem where differential expression table sorts incorrectly on click.
-
Fixed problem where very high depth samples would cause an out-of-memory error.
-
Fixed problem where mkgtf would produce incorrectly formatted GTF files.
-
Fixed problem where debug tar.gz file would be very large if the pipestance halted mid-stage.
-
Fixed problem with copying files on certain CIFS volumes.
- Initial release.