For libraries prepared with GEM-X Universal 3' Gene Expression v4 or GEM-X Universal 5’ Gene Expression v3 Reagent Kits, check the
Supported Librariestable to confirm compatibility with the latest Cell Ranger version.
Sample multiplexing is a technique that allows multiple samples to be processed together in a single experiment, significantly improving efficiency and reducing costs. By pooling samples, researchers can increase throughput while using fewer resources and less time. Sample multiplexing also reduces the risk of batch effects by ensuring that all samples are processed under the same conditions.
Unique molecular tags are assigned to each sample to enable Cell Ranger to identify and differentiate them accurately, ensuring that data is correctly matched to each sample.
10x Genomics supports the following methods of sample and cell multiplexing:
- On-chip multiplexing (OCM): Uses a microfluidic system to physically separate and process multiple samples simultaneously within the same chip, reducing sample handling complexity while maintaining high throughput.
- Hashing with Antibody Capture: Supports the use of barcoded antibodies to label individual cells from distinct samples.
- CellPlex (3' Cell Multiplexing): Uses oligonucleotide tags to label individual cells from different samples.
Some of the key advantages of sample multiplexing include:
- Increased sample throughput in a single experiment
- Increased number of cells assayed in a single experiment
- Increased number of possible replicates in a single experiment
- Detection of multiplets and their removal prior to analysis
The different multiplexing technologies and the associated Cell Ranger versions that support them are summarized in this table.
The GEM-X Universal 3'v4 and 5'v3 Gene Expression 4-plex assays provide a scalable microfluidic platform for on-chip multiplexing (OCM) of up to eight samples (two sets of up to four samples each).
Sample multiplexing is accomplished by partitioning up to four samples from each multiplexing set with corresponding gel beads (each containing a unique list of barcodes). A pool of GEMs is generated in a single recovery well for each set. The GEM pool in each recovery well is used to generate cDNA, where DNA derived from a cell shares a common barcode.

Dual indexed libraries are generated and sequenced from each pool. During data analysis, 10x Barcodes are used to computationally demultiplex the samples, ensuring accurate sample identification and separation.
Advantages of on-chip multiplexing
- Easier Multiplexing: No need to tag samples before chip loading, simplifying the process and reducing preparation time and sample loss.
- Cost Savings: Uses approximately 0.25x the reagent volume per chip layout, lowering the cost per sample.
- Flexible Approach: This method works with any sample type or assay, providing broad applicability across different experimental designs.
- Lower cell input: Supports lower cell input levels compared to CellPlex, making it suitable even when only small cell quantities are available.
Cell Ranger v9.0 and later can be used to analyze libraries generated using the GEM-X Universal 3' and 5’ Multiplex assays. Learn how to set your cellranger multi analysis for on-chip multiplexing libraries.
In this sample multiplexing technique, cells or nuclei labeled with antibody-based hashtag oligos (HTOs) are pooled before they are loaded onto a 10x Genomics chip. cDNA from poly-adenylated mRNA and DNA from the Feature Barcode are generated simultaneously from the same single cells within a Gel Bead-in-Emulsion (GEM).
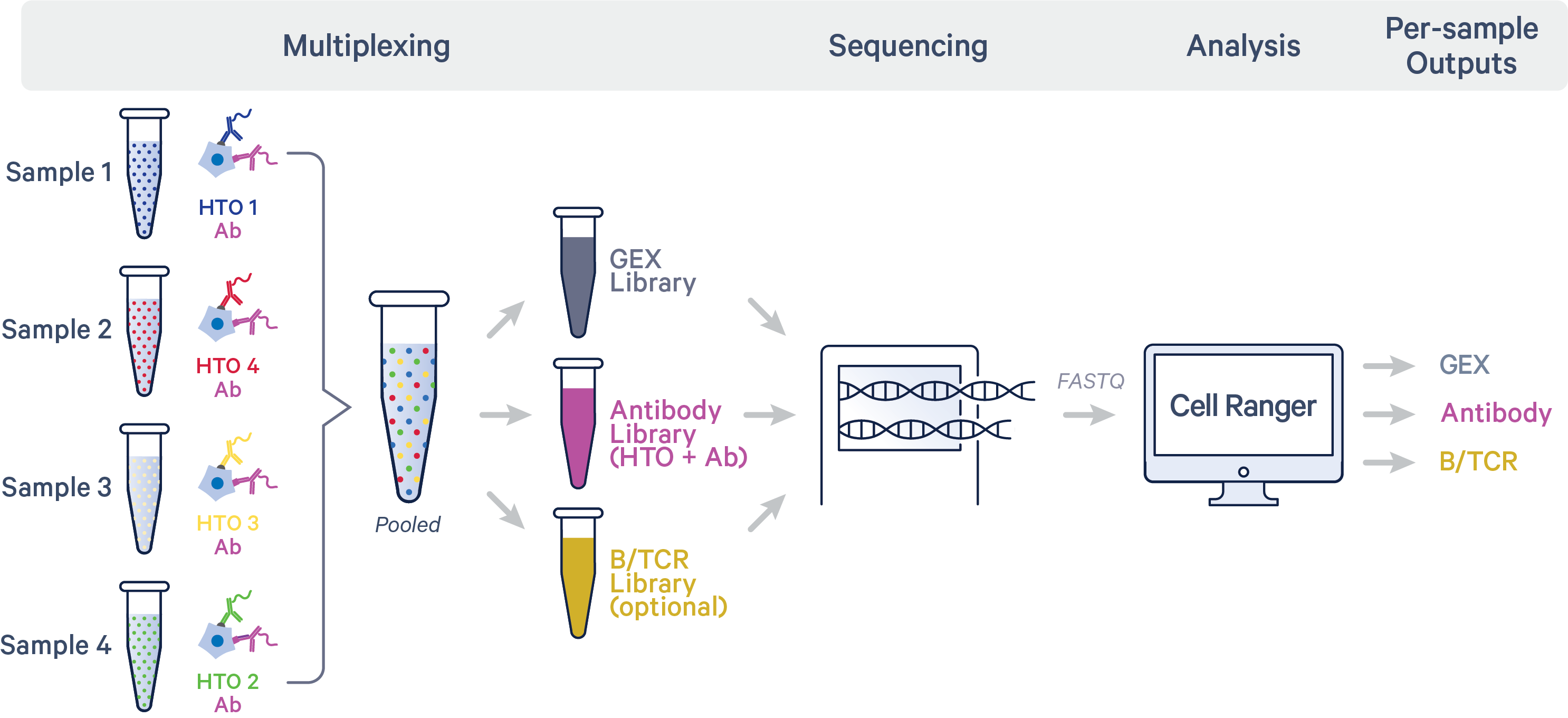
These articles provide guidance on hashing with Antibody Capture:
- Can I perform cell hashing in a GEM-X Universal 3’ Gene Expression v4 workflow?
- Can I perform cell hashing in a GEM-X Universal 5’ Gene Expression v3 workflow?
- Can I perform cell hashing in Next GEM Universal 3’ or 5’ workflows?
Starting from Cell Ranger v9.0, the analysis pipeline for hashing with Antibody Capture has been simplified. This technique is compatible with TotalSeq™-A, TotalSeq™-B, TotalSeq™-C, and Proteintech Genomics (PTG)-derived antibodies.
Libraries hashed with Antibody Capture can be analyzed using cellranger multi. To set up the required input CSV files, refer to the following guides:
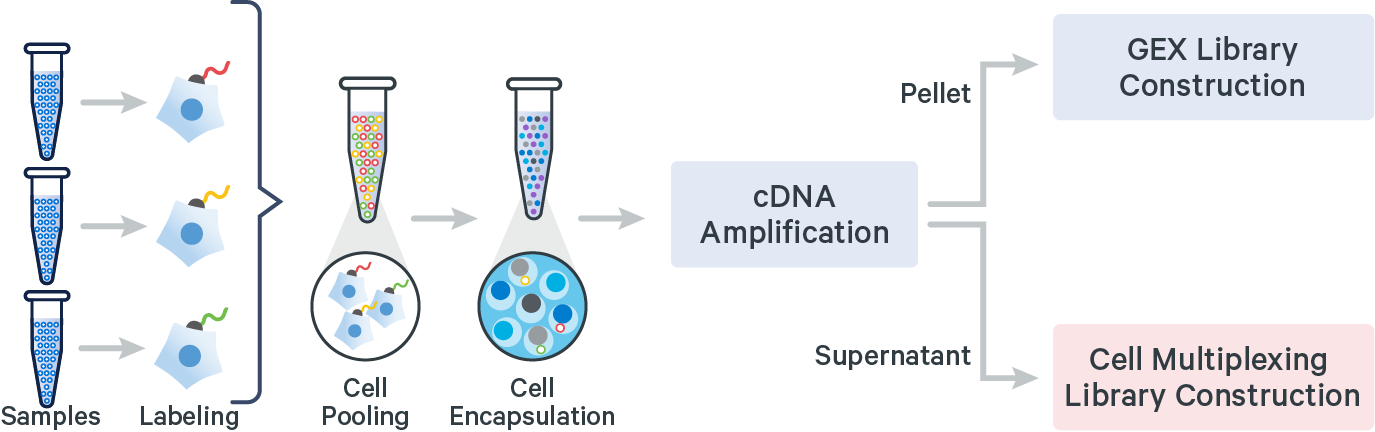
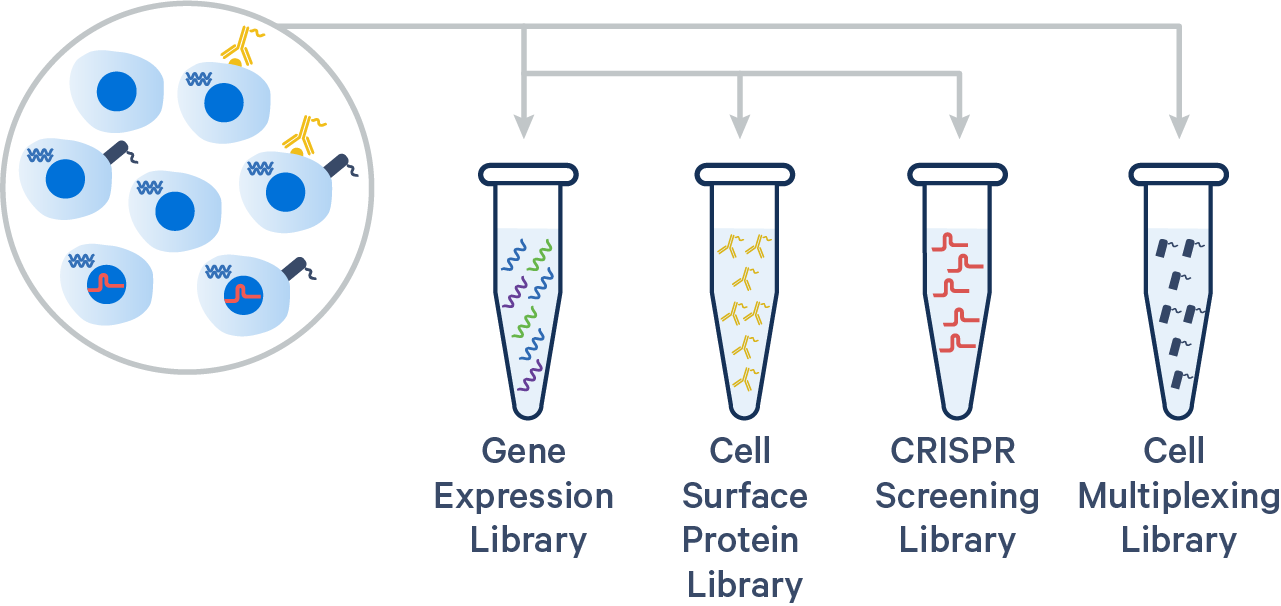
The 10x Genomics 3’ CellPlex Multiplexing Solution is a Feature Barcode technology, similar to existing 10x Genomics Cell Surface Protein and CRISPR Screening assays. Single Cell 3’ v3.1 Gel Beads and reagents can be used for CellPlex. Cells or nuclei labeled with Cell Multiplexing Oligos (CMOs) are pooled prior to loading onto a 10x Genomics chip. cDNA from poly-adenylated mRNA and DNA from the CMO Feature Barcode are generated simultaneously from the same single cells inside of a GEM. 3’ CellPlex reagents are compatible with samples whose cell surface proteins have been labeled with TotalSeq™-B antibody-oligonucleotide conjugates or samples transduced with Feature Barcode technology compatible sgRNA constructs.
Analyzing 3’ CellPlex data with Cell Ranger
Cell Ranger v6.0 or later is required to analyze Cell Multiplexing data. Multiple samples are uniquely tagged with CMOs prior to pooling in a single GEM well, resulting in a CMO and Gene Expression (GEX) library for each GEM well. After demultiplexing the BCL files, run the cellranger multi pipeline on the combined FASTQ data for the CMO and GEX libraries to obtain separate per-sample output files for each CMO.
