Cell Ranger 7.0 introduces support for Flex libraries using the cellranger multi pipeline. While some steps are similar to the existing algorithm for Gene Expression, there are a few differences illustrated and described below for singleplex and multiplex Flex experiments.
Singleplex Flex steps
Multiplex Flex steps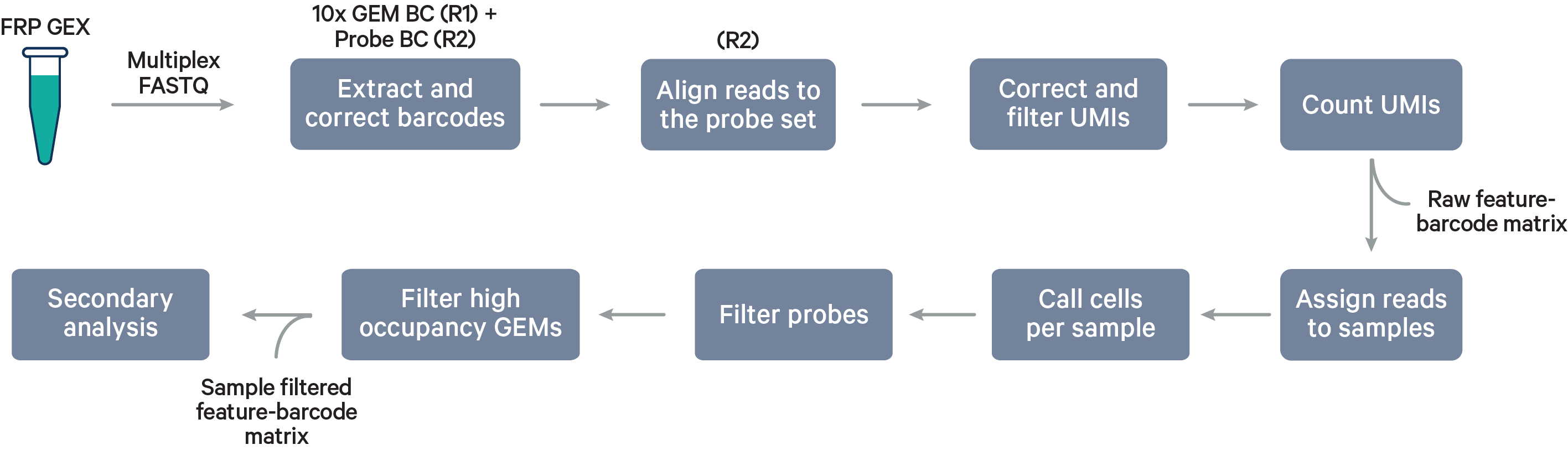
Singleplex workflow
The cell barcode will be composed only of the 10x GEM Barcode and all reads with valid barcodes are considered to be part of the sample. This configuration does not use a [samples] section in the multi config file.
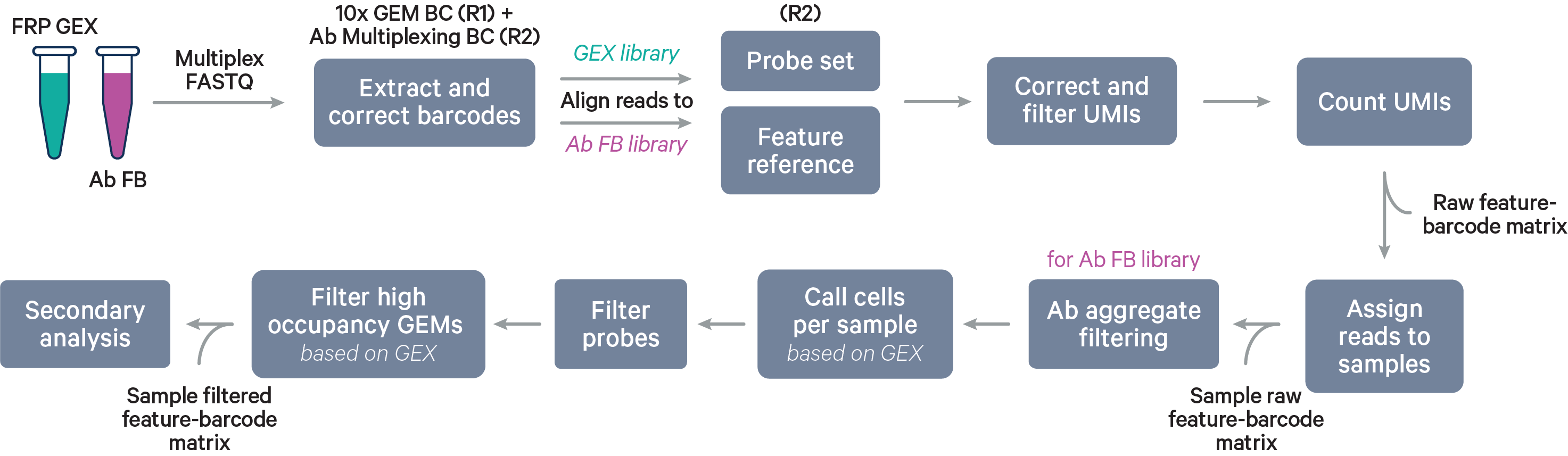
Multiplex workflow
The cell barcode will be composed of both the 10x GEM Barcode and the Probe Barcode. Reads must have valid 10x GEM and Probe Barcodes in order to be assigned to a sample. The corrected Probe Barcode sequence is mapped to its corresponding Probe Barcode ID. If this ID has been specified within the probe_barcode_ids column of the [samples] section, the read is assigned to the sample associated with this Probe Barcode ID.
Note that each Probe Barcode ID (e.g., BC001) is actually a mix of four distinct barcode sequences, which ensures balanced base composition during sequencing. Raw Probe Barcode sequences are first corrected independently of the corresponding 10x GEM Barcode using the full list of all Probe Barcode sequences from probe construction. These corrected sequences are then collapsed such that the Probe Barcode sequence used within the cell barcode is always lexicographically the first barcode amongst the four barcodes from a given mixture. For example, if the four Probe Barcodes are ACTTTAGG, CGAGGGTA, GACACTAC, and TTGCACCT, all instances of these Probe Barcodes will be converted to ACTTTAGG for use within the Cell Ranger pipeline and outputs (e.g., feature-barcode matrix). The original sequence will be maintained in uncorrected form within the raw cell barcode tag (CR) included in the BAM file.
Cell Ranger 7.0 introduces support for analyzing fixed RNA using the cellranger multi pipeline. During the fixed RNA workflow, whole transcriptome probe panels, consisting of multiple pairs of probes for each targeted gene, are added to the tissue. These probe pairs hybridize to their target transcript and are then ligated together.
To analyze fixed RNA data, the probe-set parameter must be used to specify a probe set reference CSV file. When this parameter is used, Cell Ranger will count ligation events using the probe aligner algorithm (reads are also aligned to the reference transcriptome using STAR, but only to determine their alignment positions and CIGAR strings; STAR alignments are not used to assign reads to genes for fixed RNA data).
Distinct ligation events are counted for Flex rather than distinct transcripts. Sequencing reads are aligned to the probe set reference and assigned to the genes they target. For genes targeted by more than one probe pair, the UMI counts of all probe pairs targeting that gene are summed up and output as the UMI count for that gene in the feature-barcode matrix (for more information, see this article).
The probe alignment algorithm is similar to a seed-and-extend aligner, where each half of the read is a seed, as described in detail below:
- Build an index of the half-probe sequences in the probe set reference CSV.
- For each read, look up each read half in this index, allowing up to one mismatch and no indels.
- If both halves of the read map to the same probe ID, the read is confidently mapped with a MAPQ (mapping quality) of 255.
- If one half maps and the other half does not map, compare the sequence of the unmapped half to the expected sequence for that probe, allowing mismatches but not indels. If the match exceeds the minimum alignment score (30, scoring +1 for a match and -1 for a mismatch), the read is confidently mapped.
- If the unmapped half of the read does not match the expected sequence for that probe, the read is half-mapped with a MAPQ of 1, and does not contribute to UMI counts.
- If both halves of the probe map but to different probes, the read is ambiguously mapped with a MAPQ of 3, and does not contribute to UMI counts.
- If neither probe half aligns to the probe set reference by the probe aligner algorithm, the read is unmapped and will have MAPQ=0 (column 5). If neither probe half aligns to the probe set reference by the probe aligner algorithm and the reference transcriptome by STAR, the read will be UNMAPPED (0x4) in
FLAG(column 2) and also have MAPQ=0. - All reads with up to three mismatches are guaranteed to align.
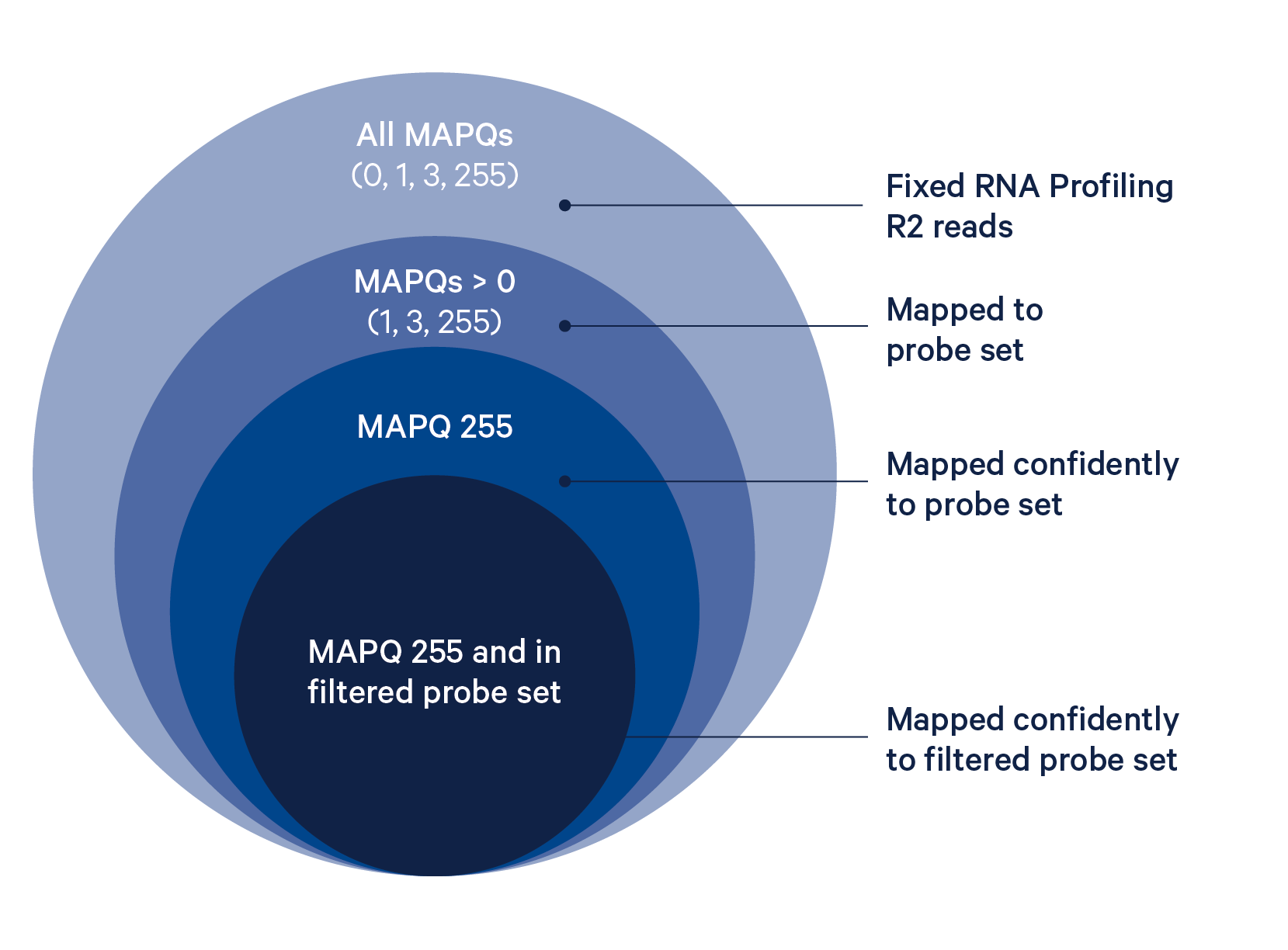
The BAM tag pr:Z reports a semicolon-separated list of probe IDs. See BAM alignment tags for a detailed description.
The cell calling algorithm is the same as described on the Gene Expression algorithm page, but done for each sample individually.
Probes that are predicted to have off-target activity to homologous genes or sequences are excluded from analysis by default (all probes are present in the raw matrix file). These probes are marked with FALSE in the included column of the probe set reference CSV. Any gene that has at least one probe with predicted off-target activity will be excluded from filtered outputs. Setting filter-probes to false in the multi config file for cellranger multi will result in UMI counts from all non-deprecated probes, including those with predicted off-target activity, to be used in the analysis. Probes whose ID is prefixed with DEPRECATED are always excluded from the analysis. Please see the Probe Set Overview for details on the probes.
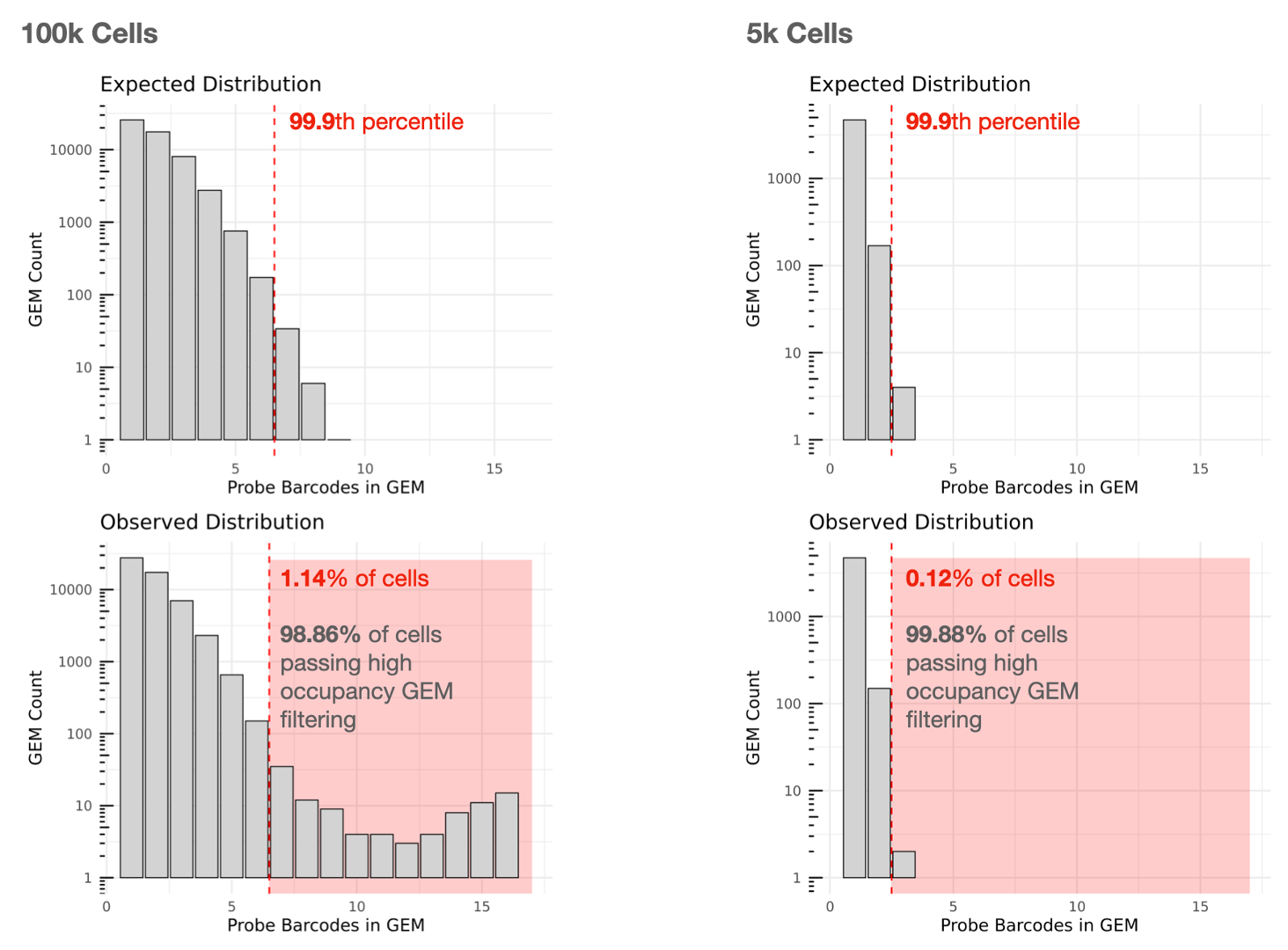
Cell calling is performed and we utilize the 10x GEM and Probe Barcodes to estimate the mean of the Poisson distribution that should describe the number of Probe Barcodes observed per GEM under optimal chip loading. Note that the mean of this distribution will increase roughly linearly with cell load. Using the expected distribution, we estimate the 99.9th percentile for the number of Probe Barcodes observed per GEM, and remove all cells belonging to GEMs that fall beyond this threshold in the observed distribution to mitigate higher than expected barcode collision rates.
High amounts of filtering due to high occupancy GEMs may reflect partial clogs/wetting failures or significant deviations from the recommended chip loading protocol. In an optimal experiment, the percentage of cells passing high occupancy GEM filtering that can be used for analysis is usually more than 90%.
The lefthand plot below shows an example of a multiplex Flex experiment with 16 Probe Barcodes aiming to capture 100,000 cells. Cells contained in GEMs falling in the red zone in the observed distribution will be removed from Cell Ranger's filtered outputs (1.14% removed, 98.86% used). Note that the Y-axes of the histograms are log10 scaled.
For comparison, the righthand plot illustrates an experiment with a lower targeted cell number (5,000 cells) with 16 Probe Barcodes, for which 0.12% of cells are removed and 99.88% can be used.
In Flex, as long as the standard workflow is followed, probe accessibility to genomic DNA (gDNA) should be limited. The expectation is that gDNA should have a negligible (less than 1%) contribution to sequenced ligation events.
The two exceptions are:
- If samples are decrosslinked during an antigen retrieval step (>70°C)
- Samples are relatively low in complexity (lower UMI counts)
Cell Ranger uses a subset of transcript probes that have a mix of exon-junction-spanning (spliced) and non-exon-junction-spanning (unspliced) probes for the same gene to estimate gDNA content. Counts from probes that do not span an exon-exon junction include some background signal, as these probes can hybridize to gDNA. Probes that do span an exon-exon junction will specifically detect spliced transcripts with no or minimal signal due to gDNA background. This method estimates the proportion of UMIs that are derived from background ligation events.
The relationship between the counts from exon-exon-junction-spanning and non-exon-junction-spanning probes can be visualized in a scatter plot as shown below.
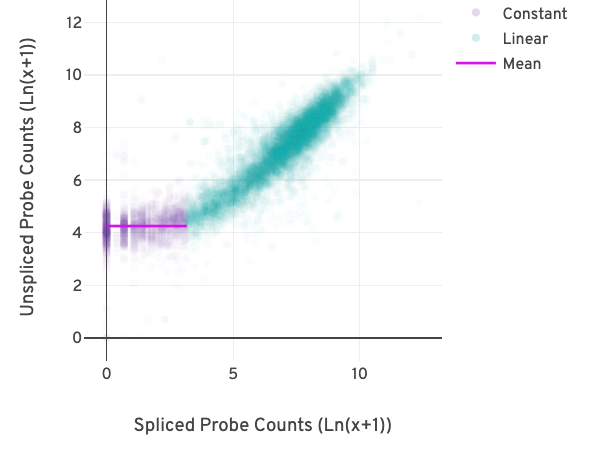
Each point in the plot reflects the relationship between two pseudo-bulk counts for a gene. The x-axis represents the sum of the junction-spanning probe counts across barcodes per gene. The y-axis represents the sum of the non-junction-spanning probe counts across barcodes for the same gene.
The difference in the counts due to background becomes apparent at low probe counts. The relationship between the pseudo-bulk counts switches from being linearly correlated (turquoise) to non-correlated (purple) in the figure above.
A segmented linear model is used to estimate where the unspliced and spliced counts begin to deviate. The mean of unspliced counts (purple) estimates the UMI background level per unspliced probe. Counts less than this value have a high probability of stemming from gDNA.
Starting in Cell Ranger 7.1, the web summary and metrics_summary.csv will report genomic DNA metrics when the v1.0.1 probe set reference CSV with the region column is used. The metrics are shown in the Library view for a singleplex experiment and in the Library and Cells views for a multiplex experiment.
-
The metric
Estimated UMIs from Genomic DNAis the estimated fraction of filtered UMIs derived from gDNA based on the discordance between probes targeting exon-junction-spanning regions and non-exon-junction-spanning regions. Normally,Estimated UMIs from Genomic DNAshould be less than 5% in Flex data with good quality samples. The fraction is calculated as follows, -
The
Estimated UMIs from genomic DNA per unspliced probeis the estimated number of UMIs derived from gDNA for each probe targeting non-exon-junction-spanning regions. A probe not spanning an exon junction with a total UMI count below this value has a high likelihood of its UMIs being derived primarily from hybridization to genomic DNA rather than mRNA.
The secondary analyses are described on the Gene Expression algorithm page.
The algorithms used for Flex experiments with Antibody Capture libraries are the same as those documented on the Antibody Capture algorithm page.
After reads are assigned to samples, the aggregate filtering algorithm is applied per sample (not per barcode), followed by per sample cell calling using the Gene Expression data. In antibody-only analyses, cell calling is done with the Antibody Capture data.
Starting in v7.2, there is a multiplexing barcode pair auto-detection algorithm for Flex multiplex GEX + Antibody Capture experiments.
The algorithm automatically detects the corresponding Antibody Multiplexing Barcode (e.g., "AB001") for a Probe Barcode (e.g., "BC001") that is specified in the multi config CSV file. If the user specifies the pairing (e.g., "BC001+AB001") in the multi config CSV file, the result of the auto-pairing algorithm is only used to provide a warning if its result disagrees with the explicit pairing.
The barcodes are paired based on an inclusion list with Probe Barcodes and Antibody Multiplexing Barcodes using a subset of all reads for each library during the chemistry detection phase. The algorithm:
- Removes any Probe Barcode (BC) or Antibody Multiplexing Barcode (AB) that appears in < 0.5% of all GEMs observed in the read subset.
- Computes the cell barcode overlap coefficient between all possible pairings of BC and AB barcodes.
- Uses a greedy approach to pair the BC and AB barcodes with the highest overlap coefficient, continuing until all multiplexing barcodes are uniquely paired or the pool of possible pairs is exhausted.
During barcode correction and feature counting, each paired AB barcode is translated into its corresponding BC barcode to ensure that gene expression and antibody feature counts from the same cell appear in the count matrix with the same cell barcode.
Barcode pair syntax:
- For a GEX + Antibody analysis, either specify all
probe_barcode_idsas "BC00X+AB00X" or "BC00X"; do not mix these formats in one config file. - For Antibody Capture-only analysis, only specify the "AB00X" barcode in the config CSV; do not mix with GEX analysis in one config file.
Barcode auto-pairing is disabled for CRISPR Guide Capture. CRISPR Multiplexing Barcodes must be specified in the multi config CSV (e.g., BC001+CR001)
For Flex multiplex GEX + Antibody Capture experiments, there is an algorithm to identify potentially unexpected correlations between pairs of BC and AB barcodes (BC-AB, BC-BC, AB-AB pairs). These correlations could be indicative of a workflow error or a mistake in the multi config CSV file if an explicit barcode pairing was provided, for example:
- One Probe Barcode is detected as being associated with more than one Antibody Multiplexing Barcode
- Antibody Multiplexing Barcode has no associated Probe Barcode
- Incorrect pairing specified in the multi config CSV (Cell Ranger will run to completion with a web summary error message about the detected incorrect pairing)
The correlations are identified using the cell barcode overlap coefficient between all possible pairings of BC and AB barcodes besides the pairs used in the analysis. Any pairing with an overlap coefficient ≥ 0.6 will be reported in the web summary. If present, barcode overlap metrics are provided in the frp_gem_barcode_overlap.csv output file. The calculation for Antibody Multiplexing Barcode contamination detection is similar to Probe Barcode contamination detection:
Overlap coefficient =