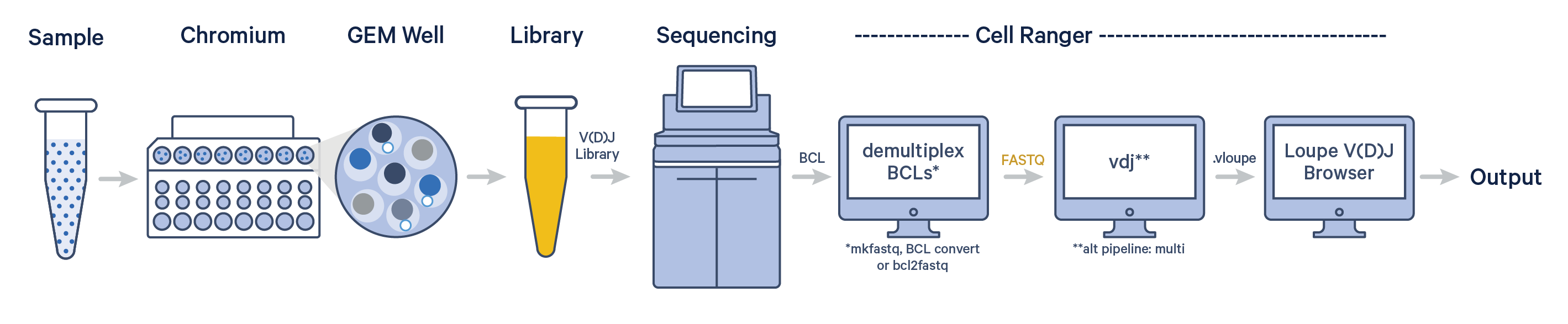
The cellranger vdj pipeline can be used to analyze sequencing data produced from Chromium Single Cell 5' V(D)J libraries. It takes FASTQ files from cellranger mkfastq, BCL Convert or bcl2fastq for V(D)J libraries and performs sequence assembly and paired clonotype calling. It uses the Chromium cellular barcodes and UMIs to assemble V(D)J transcripts per cell. Clonotypes and CDR3 sequences are output as a .vloupe file which can be loaded into Loupe V(D)J Browser.
To generate FASTQ files, refer to the instructions on running cellranger mkfastq. For help getting started, try the cellranger vdj tutorial.
For a complete list of cellranger vdj command-line arguments, run cellranger vdj --help.
To generate single cell V(D)J sequences and annotations for a single library, run cellranger vdj with these required arguments:
| Argument | Description |
|---|---|
--id | Required. A unique run ID string: e.g. sample345 |
--fastqs | Required. Path of the FASTQ folder generated by cellranger mkfastq e.g. /home/jdoe/runs/HAWT7ADXX/outs/fastq_path. Can take multiple comma-separated paths, which is helpful if the same library was sequenced on multiple flowcells. Doing this will treat all reads from the library, across flowcells, as one sample. |
--reference | Required. Path to the Cell Ranger V(D)J compatible reference e.g. /opt/refdata-cellranger-vdj-GRCh38-alts-ensembl-7.1.0. If --denovo is specified, this parameter is optional. |
--sample | Required. Sample name as specified in the sample sheet supplied to mkfastq. Can take multiple comma-separated values, which is helpful if the sample was sequenced on multiple flowcells and the sample name used (and therefore fastq file prefix) is not identical between them. Doing this will treat all reads from the library, across flowcells, as one sample. |
For a complete list of cellranger vdj command-line arguments, run cellranger vdj --help or visit the Cell Ranger Commands page.
After determining your input arguments and options, run cellranger vdj:
cd /home/jdoe/runs
cellranger vdj --id=sample345 \
--reference=/opt/refdata-cellranger-vdj-GRCh38-alts-ensembl-7.1.0 \
--fastqs=/home/jdoe/runs/HAWT7ADXX/outs/fastq_path \
--sample=mysample \
--localcores=8 \
--localmem=64
Following a set of preflight checks to validate input arguments, cellranger vdj pipeline stages will begin to run:
Martian Runtime - v4.0.8
Running preflight checks (please wait)...
yyyy-mm-dd hh:mm:ss [runtime] (ready) ID.sample345.SC_VDJ_ASSEMBLER_CS.VDJ_PREFLIGHT
yyyy-mm-dd hh:mm:ss [runtime] (run:local) ID.sample345.SC_VDJ_ASSEMBLER_CS.VDJ_PREFLIGHT.fork0.chnk0.main
yyyy-mm-dd hh:mm:ss [runtime] (ready) ID.sample345.SC_VDJ_ASSEMBLER_CS.VDJ_PREFLIGHT_LOCAL
...
By default, cellranger will use all of the cores available on your system to execute pipeline stages. You can specify a different number of cores to use with the --localcores option; for example, --localcores=16 will limit cellranger to using up to sixteen cores at once. Similarly, --localmem will restrict the amount of memory (in GB) used by cellranger.
The pipeline will create a new folder named with the sample ID you specified (e.g. /home/jdoe/runs/sample345) for its output. If this folder already exists, cellranger will assume it is an existing pipestance and attempt to resume running it.
A successful cellranger vdj run should conclude with a message similar to this:
Outputs:
- Run summary HTML: /home/jdoe/runs/sample345/outs/web_summary.html
- Run summary CSV: /home/jdoe/runs/sample345/outs/metrics_summary.csv
- Clonotype info: /home/jdoe/runs/sample345/outs/clonotypes.csv
- Filtered contig sequences FASTA: /home/jdoe/runs/sample345/outs/filtered_contig.fasta
- Filtered contig sequences FASTQ: /home/jdoe/runs/sample345/outs/filtered_contig.fastq
- Filtered contigs (CSV): /home/jdoe/runs/sample345/outs/filtered_contig_annotations.csv
- All-contig FASTA: /home/jdoe/runs/sample345/outs/all_contig.fasta
- All-contig FASTA index: /home/jdoe/runs/sample345/outs/all_contig.fasta.fai
- All-contig FASTQ: /home/jdoe/runs/sample345/outs/all_contig.fastq
- Read-contig alignments: /home/jdoe/runs/sample345/outs/all_contig.bam
- Read-contig alignment index: /home/jdoe/runs/sample345/outs/all_contig.bam.bai
- All contig annotations (JSON): /home/jdoe/runs/sample345/outs/all_contig_annotations.json
- All contig annotations (BED): /home/jdoe/runs/sample345/outs/all_contig_annotations.bed
- All contig annotations (CSV): /home/jdoe/runs/sample345/outs/all_contig_annotations.csv
- Barcodes that are declared to be targetted cells: /home/jdoe/runs/sample345/outs/cell_barcodes.json
- Clonotype consensus FASTA: /home/jdoe/runs/sample345/outs/consensus.fasta
- Clonotype consensus FASTA index: /home/jdoe/runs/sample345/outs/consensus.fasta.fai
- Contig-consensus alignments: /home/jdoe/runs/sample345/outs/consensus.bam
- Contig-consensus alignment index: /home/jdoe/runs/sample345/outs/consensus.bam.bai
- Clonotype consensus annotations (CSV): /home/jdoe/runs/sample345/outs/consensus_annotations.csv
- Concatenated reference sequences: /home/jdoe/runs/sample345/outs/concat_ref.fasta
- Concatenated reference index: /home/jdoe/runs/sample345/outs/concat_ref.fasta.fai
- Contig-reference alignments: /home/jdoe/runs/sample345/outs/concat_ref.bam
- Contig-reference alignment index: /home/jdoe/runs/sample345/outs/concat_ref.bam.bai
- Loupe V(D)J Browser file: /home/jdoe/runs/sample345/outs/vloupe.vloupe
- V(D)J reference:
fasta:
regions: /home/jdoe/runs/sample345/outs/vdj_reference/fasta/regions.fa
donor_regions: /home/jdoe/runs/sample345/outs/vdj_reference/fasta/donor_regions.fa
reference: /home/jdoe/runs/sample345/outs/vdj_reference/reference.json
- AIRR Rearrangement TSV: /home/jdoe/runs/sample345/outs/airr_rearrangement.tsv
- All contig info (ProtoBuf format): /home/jdoe/runs/sample345/outs/vdj_contig_info.pb
Waiting 6 seconds for UI to do final refresh.
Pipestance completed successfully!
The output folder name is the same as the sample ID you specified (e.g. sample345). The outs subfolder contains the main pipeline output files.
Once cellranger vdj has successfully completed, you can browse the resulting summary HTML file in any supported web browser, open the .vloupe file in Loupe V(D)J Browser, or refer to the Understanding Output section to explore the data by hand.