The Chromium Single Cell 5’ Barcode Enabled Antigen Mapping (BEAM) technology offers a scalable approach for mapping a V(D)J receptor to a target antigen by enabling the detection of gene expression profiles, paired V(D)J receptors, and signal from a bound antigen from the same single cell. All of these libraries, generated from a single GEM well, can be analyzed together with Cell Ranger v7.1 or later using the cellranger multi pipeline.
Hereafter libraries created using the BEAM-T workflow are called TCR Antigen Capture libraries, whereas libraries created using the BEAM-Ab workflow are called BCR Antigen Capture libraries. Antigen Capture without a prefix refers to either TCR or BCR Antigen Capture library, depending on the context.
To analyze an antigen library created using an antigen-multimer staining assay (TotalSeq™-C, Immudex's dMHC Dextramer® libraries with dCODE Dextramers), visit the TotalSeq™-C section of the 3' Chromium Single Cell Gene Expression with Feature Barcode technology page.
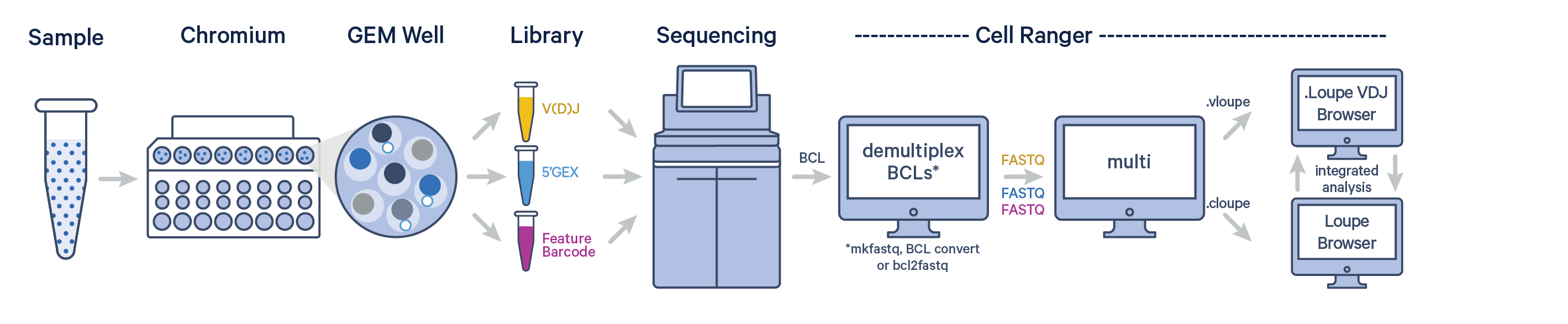
Antigen Capture + VDJ + Gene Expression (GEX) is the minimum set of libraries necessary to process antigen data from a single experiment. Here is a list of supported and unsupported library combinations. Please note that this table is not an exhaustive list.
| Library Combination | Supported? |
|---|---|
| TCR Antigen Capture + VDJ-T + GEX | Yes |
| BCR Antigen Capture + VDJ-B + GEX | Yes |
| Antigen Capture + VDJ + GEX + Antibody Capture | Yes |
| TCR Antigen Capture + VDJ-T-GD + GEX | Allowed but unsupported |
| Antigen Capture only | No |
| Antigen Capture + VDJ | No |
| TCR Antigen Capture + BCR Antigen Capture | No |
| Antigen Capture + CRISPR Guide Capture | No |
The following inputs are needed to analyze Antigen Capture, V(D)J, and Gene Expression libraries together using the cellranger multi pipeline:
Any Cell Ranger workflow starts by demultiplexing the Illumina sequencer's base call files (BCLs) for each flow cell directory into FASTQ files. Follow the instructions on running cellranger mkfastq, Illumina's BCL convert, or bcl2fastq to generate Gene Expression, V(D)J, and Antigen Capture FASTQ files. If you are already starting with FASTQ files, you can skip this step.
A Feature Reference CSV file is required when processing any Feature Barcode data. General details of this file are available on the 3' Single Cell Gene Expression website.
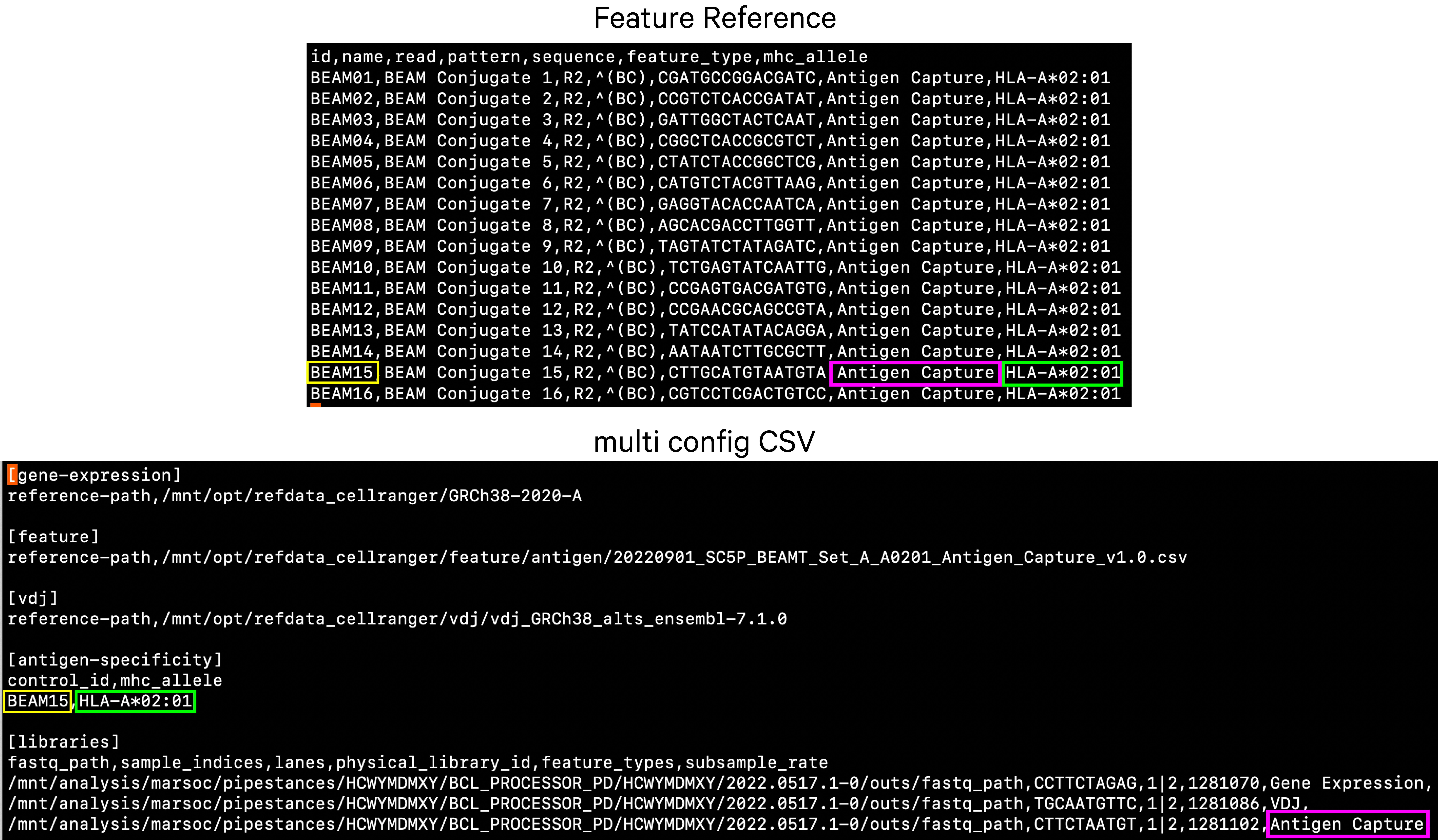
If a TCR Antigen Capture library is present, the Feature Reference must have an additional column, mhc_allele, that defines the MHC allele associated with each antigen included in the experiment.
id,name,read,pattern,sequence,feature_types,mhc_allele
ag1,ag1,R2,^(BC),CGATGCCGGACGATC,Antigen Capture,A*01
ag2,ag2,R2,^(BC),CCGTCTCACCGATAT,Antigen Capture,A*01
neg_control01,neg_control01,R2,^(BC),GATTGGCTACTCAAT,Antigen Capture,A*01
ag4,ag4,R2,^(BC),CGGCTCACCGCGTCT,Antigen Capture,A*02
ag5,ag5,R2,^(BC),CTATCTACCGGCTCG,Antigen Capture,A*02
neg_control02,neg_control02,R2,^(BC),CATGTCTACGTTAAG,Antigen Capture,A*02
ab1,ab1,R2,^(BC),GAGGTACACCAATCA,Antibody Capture,A*01
If one Antigen Capture id has an entry in the mhc_allele column, all IDs must have an entry in that column. The mhc_allele name for the negative control peptide must match the name specified in the [antigen-specifity] section of the multi config CSV. Similarly, the id field for the control antigen in the Feature Reference CSV must match the control_id in the multi config CSV.
For BCR Antigen Capture, the mhc_allele column must NOT be included:
id,name,read,pattern,sequence,feature_types
ag1,ag1,R2,^(BC),CGATGCCGGACGATC,Antigen Capture
ag2,ag2,R2,^(BC),CCGTCTCACCGATAT,Antigen Capture
ag4,ag4,R2,^(BC),CGGCTCACCGCGTCT,Antigen Capture
ag5,ag5,R2,^(BC),CTATCTACCGGCTCG,Antigen Capture
ag6,ag6,R2,^(BC),CATGTCTACGTTAAG,Antigen Capture
ag7,ag7,R2,^(BC),GAGGTACACCAATCA,Antigen Capture
ag8,ag8,R2,^(BC),AGCACGACCTTGGTT,Antigen Capture
neg_control01,neg_control01,R2,^(BC),GATTGGCTACTCAAT,Antigen Capture
The id field for the control antigen in the Feature Reference CSV must match the control_id in the multi config CSV.
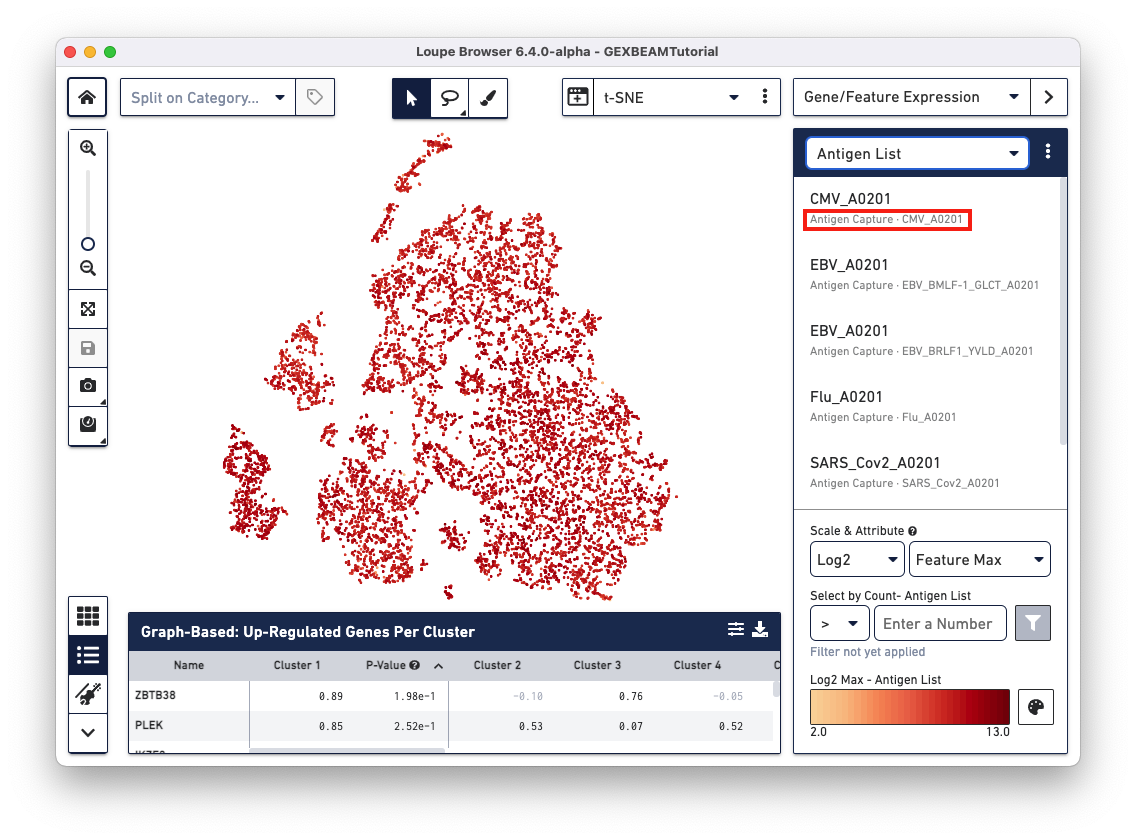
Loupe V(D)J Browser v5.0 and Loupe Browser v6.4 extract antigen names from the Feature Reference CSV. These names cannot be changed via the browser interface. In this Feature Reference example, the id and name fields have the same entry for each antigen. In Loupe Browser, the name field corresponds to the antigen name in grey (as shown below):
The Feature Barcode sequences of available BEAM conjugates section lists all the BEAM conjugate Feature Barcode sequences.
| BEAM Conjugate | Sequence |
|---|---|
| BEAM Conjugate 1 | CGATGCCGGACGATC |
| BEAM Conjugate 2 | CCGTCTCACCGATAT |
| BEAM Conjugate 3 | GATTGGCTACTCAAT |
| BEAM Conjugate 4 | CGGCTCACCGCGTCT |
| BEAM Conjugate 5 | CTATCTACCGGCTCG |
| BEAM Conjugate 6 | CATGTCTACGTTAAG |
| BEAM Conjugate 7 | GAGGTACACCAATCA |
| BEAM Conjugate 8 | AGCACGACCTTGGTT |
| BEAM Conjugate 9 | TAGTATCTATAGATC |
| BEAM Conjugate 10 | TCTGAGTATCAATTG |
| BEAM Conjugate 11 | CCGAGTGACGATGTG |
| BEAM Conjugate 12 | CCGAACGCAGCCGTA |
| BEAM Conjugate 13 | TATCCATATACAGGA |
| BEAM Conjugate 14 | AATAATCTTGCGCTT |
| BEAM Conjugate 15 | CTTGCATGTAATGTA |
| BEAM Conjugate 16 | CGTCCTCGACTGTCC |
The cellranger multi pipeline requires a configuration CSV file as input, referred to here as the multi config CSV. The multi config CSV contains paths to FASTQ files for V(D)J, Gene Expression, and Antibody Capture (BEAM) libraries, as well as a path to the Feature Reference CSV.
Main sections in a multi config CSV:
The multi config CSV contains both the library definitions and experiment configuration variables, comprising up to five sections: - [gene-expression] - [feature] - [vdj] - [antigen-specificity] - [libraries]
Go to the Cell Ranger Multi Config CSV page for a complete list of options for each section.
Go to the Cell Ranger Multi Config CSV page for a complete list of options for each section.
Example multi config CSVs can be downloaded from public datasets. Cell Ranger v7.1 and later also provides the option to download a multi config CSV template via the command line.
Here are the example multi config CSVs for a few commonly used library combinations with Antigen Capture. Make sure to replace /path/to with the full path to your data, and customize the experiment's sample/library/file names.
[gene-expression]
reference,/path/to/transcriptome
[vdj]
reference,/path/to/vdj_reference
[feature]
reference,/path/to/feature_ref.csv
[antigen-specificity]
control_id,
neg_control,
[libraries]
fastq_id,fastqs,lanes,feature_types,subsample_rate
GEX_fastqs_id,/path/to/GEX_fastqs,1|2,Gene Expression,
VDJ_B_fastqs_id,/path/to/vdj_B_fastqs,1|2,VDJ-B,
BEAM_fastqs_id,/path/to/Ag_fastqs,1|2,Antigen Capture,
[gene-expression]
reference,/path/to/transcriptome
[vdj]
reference,/path/to/vdj_reference
[feature]
reference,/path/to/feature_ref.csv
[antigen-specificity]
control_id,mhc_allele
neg_control01,A*01
neg_control02,A*02
[libraries]
fastq_id,fastqs,lanes,feature_types,subsample_rate
GEX_fastqs_id,/path/to/GEX_fastqs,1|2,Gene Expression,
VDJ_T_fastqs_id,/path/to/vdj_B_fastqs,1|2,VDJ-T,
BEAM_fastqs_id,/path/to/Ag_fastqs,1|2,Antigen Capture,
[gene-expression]
reference,/path/to/transcriptome
[vdj]
reference,/path/to/vdj_reference
[feature]
reference,/path/to/feature_ref.csv
[antigen-specificity]
control_id,mhc_allele
neg_control01,A*01
neg_control02,A*02
[libraries]
fastq_id,fastqs,lanes,feature_types,subsample_rate
GEX_fastqs_id,/path/to/GEX_fastqs,1|2,Gene Expression,
VDJ_B_fastqs_id,/path/to/vdj_B_fastqs,1|2,VDJ-B,
BEAM_fastqs_id,/path/to/BEAM_fastqs,1|2,Antigen Capture,
Antibody_fastqs_id,/path/to/AB_fastqs,1|2,Antibody Capture,
Generate a multi config CSV template by running cellranger multi-template, see usage here.
These images highlight the relationship between a TCR Antigen Capture Feature Reference CSV and its multi config CSV:
Go back to the Cell Ranger multi page to learn more about running cellranger multi.