Cloud Analysis introduces a new pipeline for cell type annotation, which can be applied to standard Cell Ranger count and multi outputs to generate accurate cell type labels. The cell annotation models are currently under beta development.
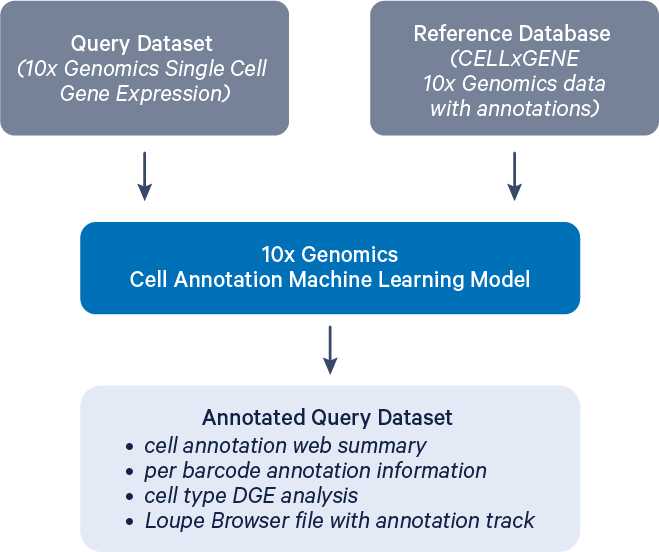
This page explains how to run the annotate pipeline and outlines the expected output files using a publicly available dataset.
Prerequisites
- A successfully completed run of either the
cellranger countormultipipeline on Cloud Analysis, with a dataset that includes a Gene Expression library. - The sample must be human (or human-derived) or mouse (or mouse-derived).
- The analysis supports datasets ranging from 100 to 800,000 cells.
- The availability of automated cell annotation is subject to restrictions based on U.S. or local laws and regulations. See regional restrictions for the list of impacted regions.
Start with the outputs of cellranger count or cellranger multi:
Before running the cell type annotation, ensure that you have already processed your data using either the cellranger count or cellranger multi pipeline. These pipelines must be run in Cloud Analysis, as you cannot begin by simply uploading the outputs.
Note: A .cloupe file will only be generated if secondary analysis is enabled during the initial run.
Go to the Analysis tab in the interface and select the analysis you wish to annotate. Click the Run Cell Type Annotation button to start setting up.
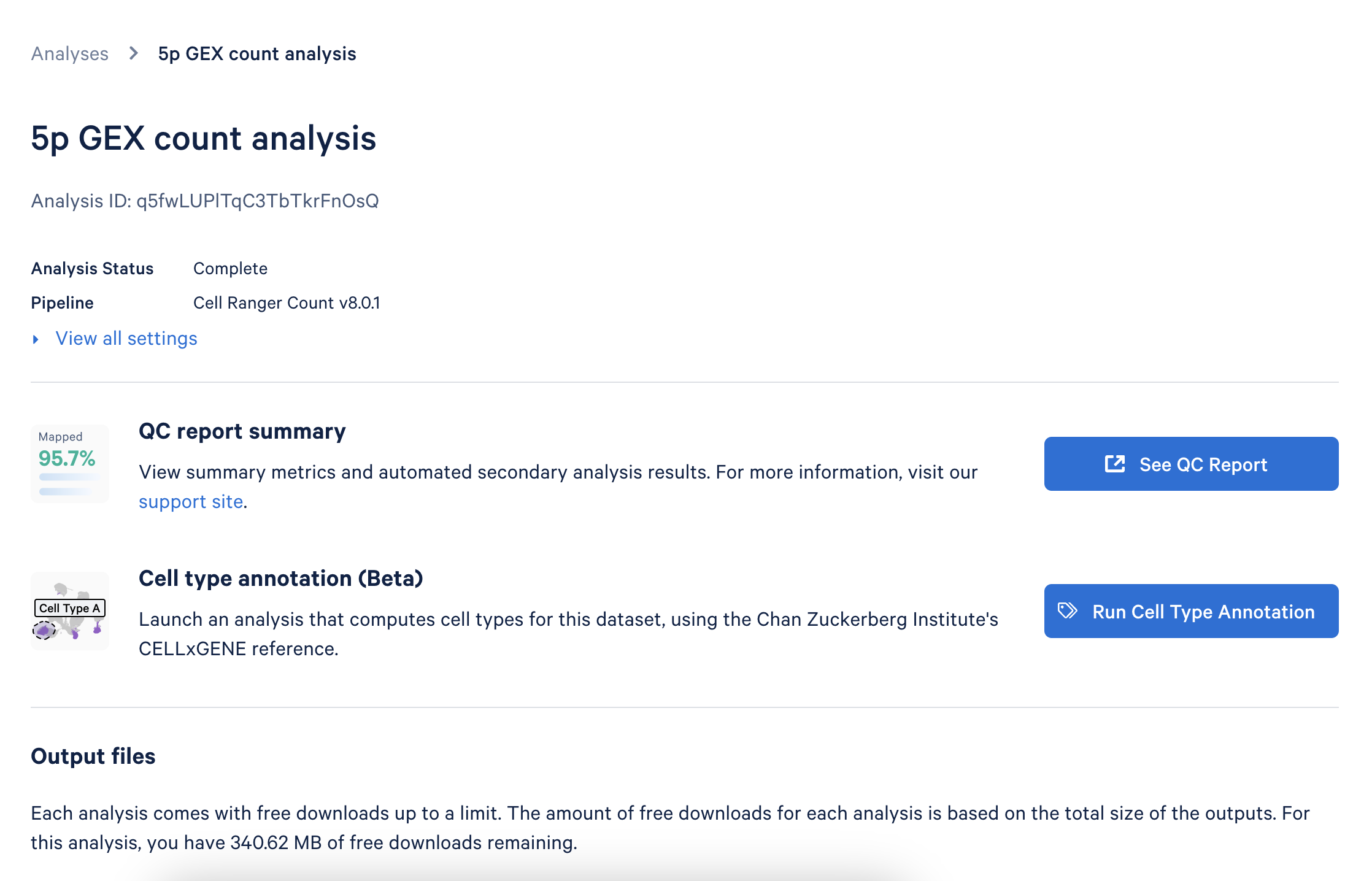
This will take you to the annotation setup page:
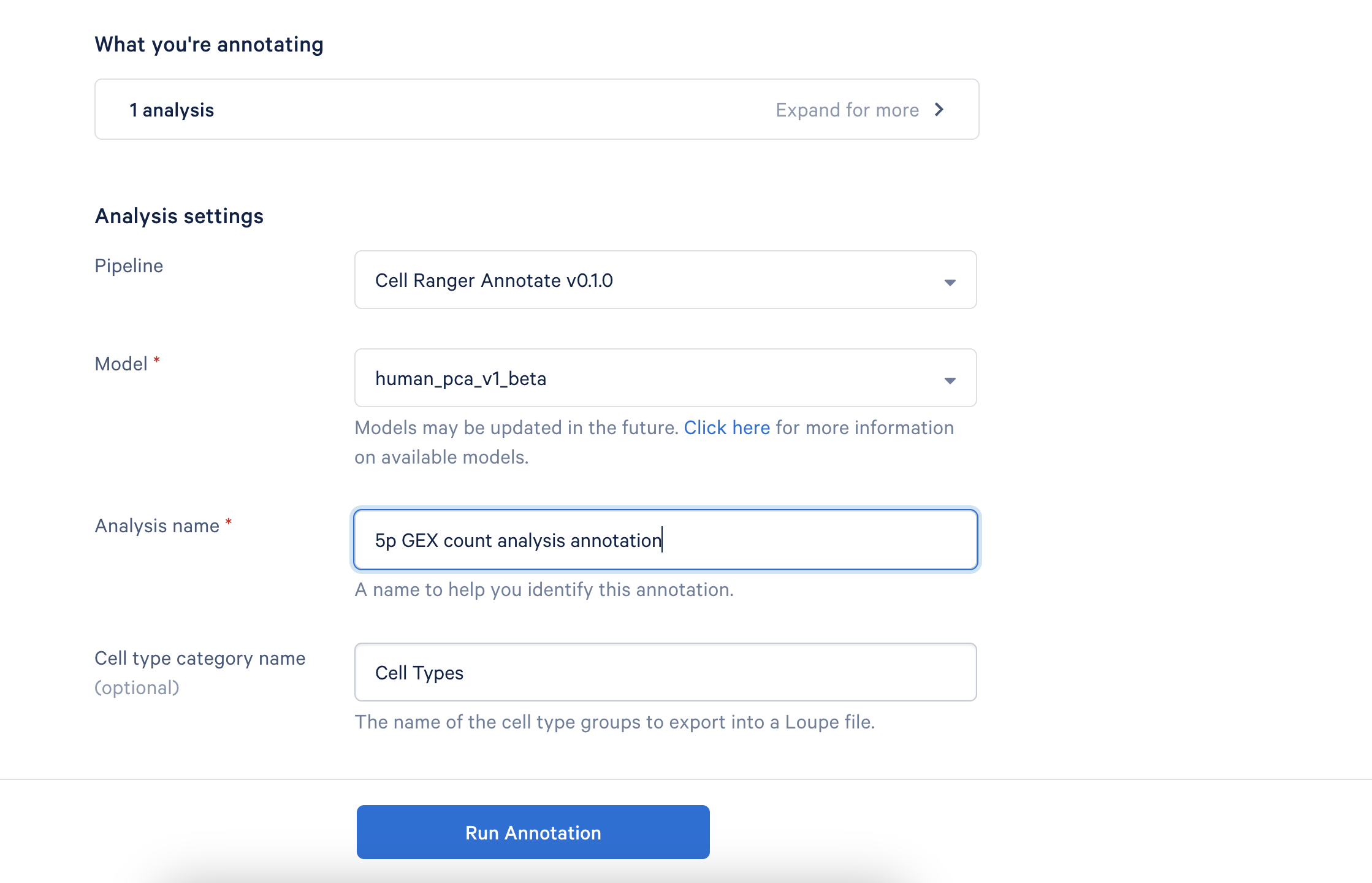
Provide a descriptive name for your analysis to help you easily identify it later. If desired, you can rename the "Cell Type Category" that appears in Loupe Browser.
There are no tunable parameters in the cell annotation algorithm at this time. As additional models are released, you will be able to select your preferred model, but no further customization options will be available.
Click Run Annotation to begin the process. You will receive an email notification once the analysis is complete.
| File | Downloaded file name | Description |
|---|---|---|
| Annotation web summary | web_summary_cell_types.html | View high-level cell types, metrics, and distribution. |
| Loupe Browser file | cell_annotation_sample_cloupe.cloupe | The Loupe Browser file from the original analysis, annotated with high-level cell types. |
| Annotation by cell | cell_annotation_results.json.gz | Detailed evidence of how each cell has been assigned a cell type by the algorithm, broken down by dataset IDs in the reference database and nearest-neighbors in each. |
| Cell types CSV file | cell_types.csv | A CSV file listing course and fine cell types for each cell. |
| Differential expression CSV | cell_annotation_differential_expression.csv | Table listing genes that are differentially expressed in each detected cell type, along with log2 fold-change and associated p-value. |
Descriptions of the output files are available on the Cell Ranger support website.