Cell annotations refer to the process of categorizing and assigning cell types to individual cells based on their gene expression profiles. These annotations are needed for understanding the cellular composition and diversity within a sample.
Cell Ranger v9.0 introduces support for cell type annotation as part of cellranger multi and cellranger count commands and as a standalone command called cellranger annotate.
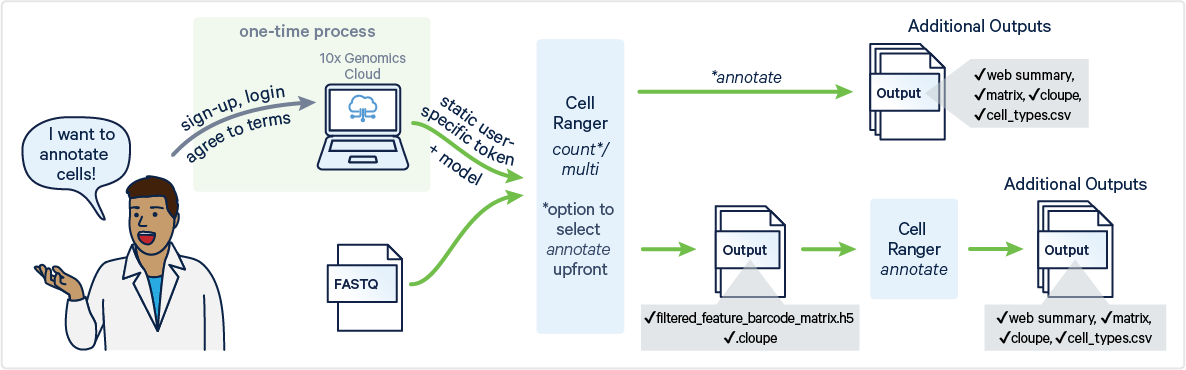
The pipeline assigns cell types by comparing each cell's gene expression profile to annotated reference datasets, rather than relying on known marker genes or tissue-specific references. Cell Ranger's annotation feature provides a starting point for annotating your samples. For more refined annotations, you can investigate specific marker genes.
Specifically, each cell barcode's gene expression data is compared to the Chan Zuckerberg CELL by GENE (CZ CELLxGENE) census, identifying the most similar cell types. A consensus label is then assigned to each barcode, with the results summarized in the web_summary.html. These labels can be viewed in Loupe Browser or accessed via the cell_types.csv output file.
Learn more about
| Pipelines | Annotation Setup | Supported Library Types for Annotation |
|---|---|---|
cellranger annotate | Compatible with outputs of cellranger multicellranger countcellranger aggr Point to filtered_feature_bc_matrix.h5 and optional .cloupe file | |
cellranger count | Enable with a parameter in the command for count | |
cellranger multi | Enable by specifying it in the multi_config.csv |
*Multiome GEX is enabled but unsupported
Cell annotation is not available for Space Ranger outputs.
Cell annotation is a complex challenge that lacks a universal solution. While we have rigorously benchmarked our method across various tissue types, the diversity of tissues and cell states (e.g., cancerous, diseased, juvenile, embryonic) far exceeds what we can comprehensively test in-house. As a result, the annotation pipeline has not undergone the same level of testing as our other algorithms. We are releasing the annotate pipeline as a beta feature with the intent to collect feedback.
We encourage customers to use this tool as a starting point for identifying cell types of interest, aiming to streamline a process that is typically manual and time-consuming. This beta release helps us gather meaningful feedback from the community so that we can continue to improve the models. Please note that annotate may remain in beta, depending on the scope and nature of feedback we receive.
The cell annotation pipeline currently supports annotations for human (or human-derived) and mouse (or mouse-derived) samples only; custom references are not yet supported.
The algorithm will ignore the presence of additional genes (e.g., a reference modified to included GFP) and will run successfully as long as the majority of genes originate from a 10x reference.
Barnyard samples are not supported.