Simplifying spatial transcriptomics protocols with Visium CytAssist
Spatial transcriptomics, commercialized as Visium Spatial Gene Expression, is a molecular profiling method that enables scientists to map whole transcriptome gene activity across an entire tissue section alongside H&E morphological imaging.
The next evolution of this platform—powered by Visium CytAssist, a benchtop instrument that facilitates the transfer of probes from tissues on standard glass slides to Visium slides—enables truly unbiased spatial discovery across an expanding range of compatible tissues with maximal experimental efficiency.
We introduced some of the advantages of Visium CytAssist in our recent webinar, including a comparison with the direct placement approach, an overview of CytAssist-compatible species and tissue conditions, workflow details, data QC metrics, the Visium CytAssist product roadmap, plus a helpful Q&A session. Review the recap in this blog, or watch the webinar for yourself here →
Visium CytAssist is also an integral part of the Visium HD workflow, together enabling whole transcriptome spatial discovery at single cell scale with continuous tissue coverage and precise transcript localization in even the smallest anatomical structures. Learn about Visium HD and the role CytAssist plays here →
How does Visium CytAssist make spatial sample management easier?
Only a few, technically-skilled, highly detail-oriented researchers can say that their favorite lab task is cryosectioning. Tissue and slide handling can be one of the more difficult aspects of spatial transcriptomics experiments, as it brings together a skillset that may be unfamiliar to scientists who haven’t worked with tissue before. That’s exactly why we developed Visium CytAssist, so that you can be the biological expert of your spatial experiments even if you’re not a histological expert.
One of the biggest CytAssist game-changers is the ability to section tissue directly onto glass slides. That means the Visium CytAssist workflow follows a straightforward, standard histological workflow: sectioning, tissue preparation, staining (H&E or IF), and imaging all take place on a standard glass slide.
After some additional tissue preparation steps, probe hybridization takes place on the glass slide too. In this step, probes hybridize to ~18,000 genes, or RNA targets, within the tissue section to achieve whole transcriptome gene expression profiling. You can then place two standard glass slides and a Visium slide with two Capture Areas into the CytAssist instrument so that the tissue sections on the standard slides can be aligned on top of the Capture Areas. Within the instrument, a brightfield image is captured to provide spatial orientation for data analysis, followed by hybridization of transcriptomic probes from the tissue onto the Visium slide. The remaining steps, starting with probe extension, follow the standard Visium workflow outside of the instrument.
Find detailed video demonstrations for the Visium CytAssist Spatial Gene Expression workflow, including instructions for FFPE tissue preparation, deparaffinization, staining, and imaging, with our how-to video series →
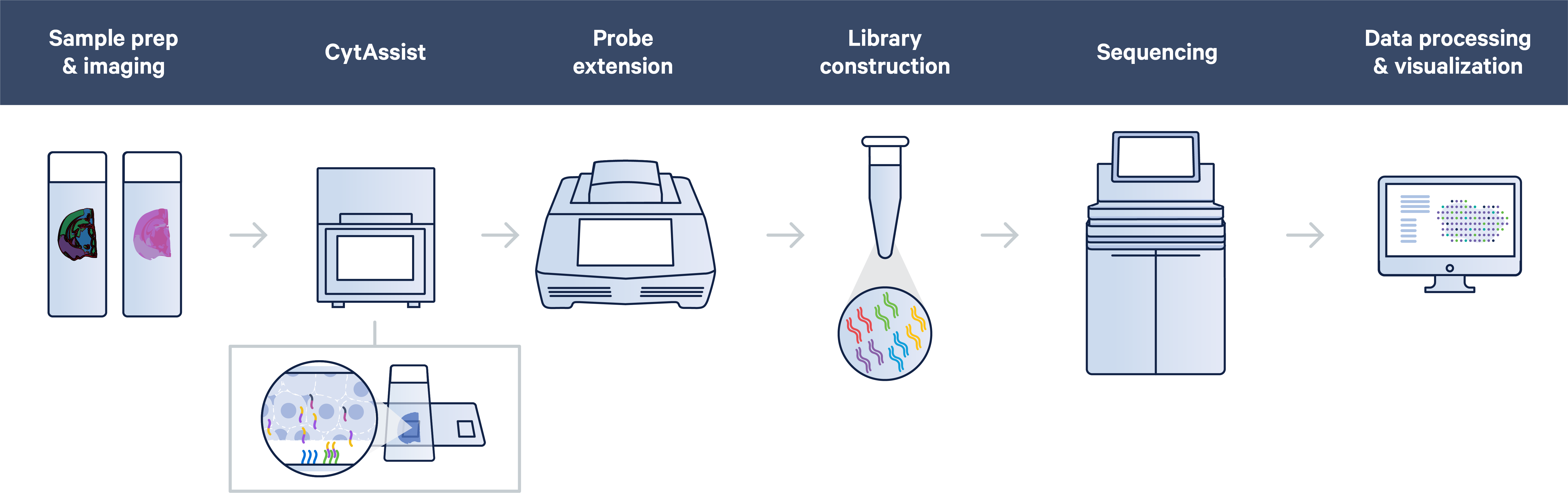

This combination of mechanical and chemical processes facilitates the transfer of transcriptomic probes from standard glass slides to Visium slides, streamlining sample management by allowing you to section tissue directly onto glass slides or start your spatial experiments with pre-sectioned tissue slides.
Additionally, the CytAsisst instrument allows you to both pre-screen your tissue sections and orient tissue sections to align the most desired region of tissue onto the Capture Area of the Visium slide.
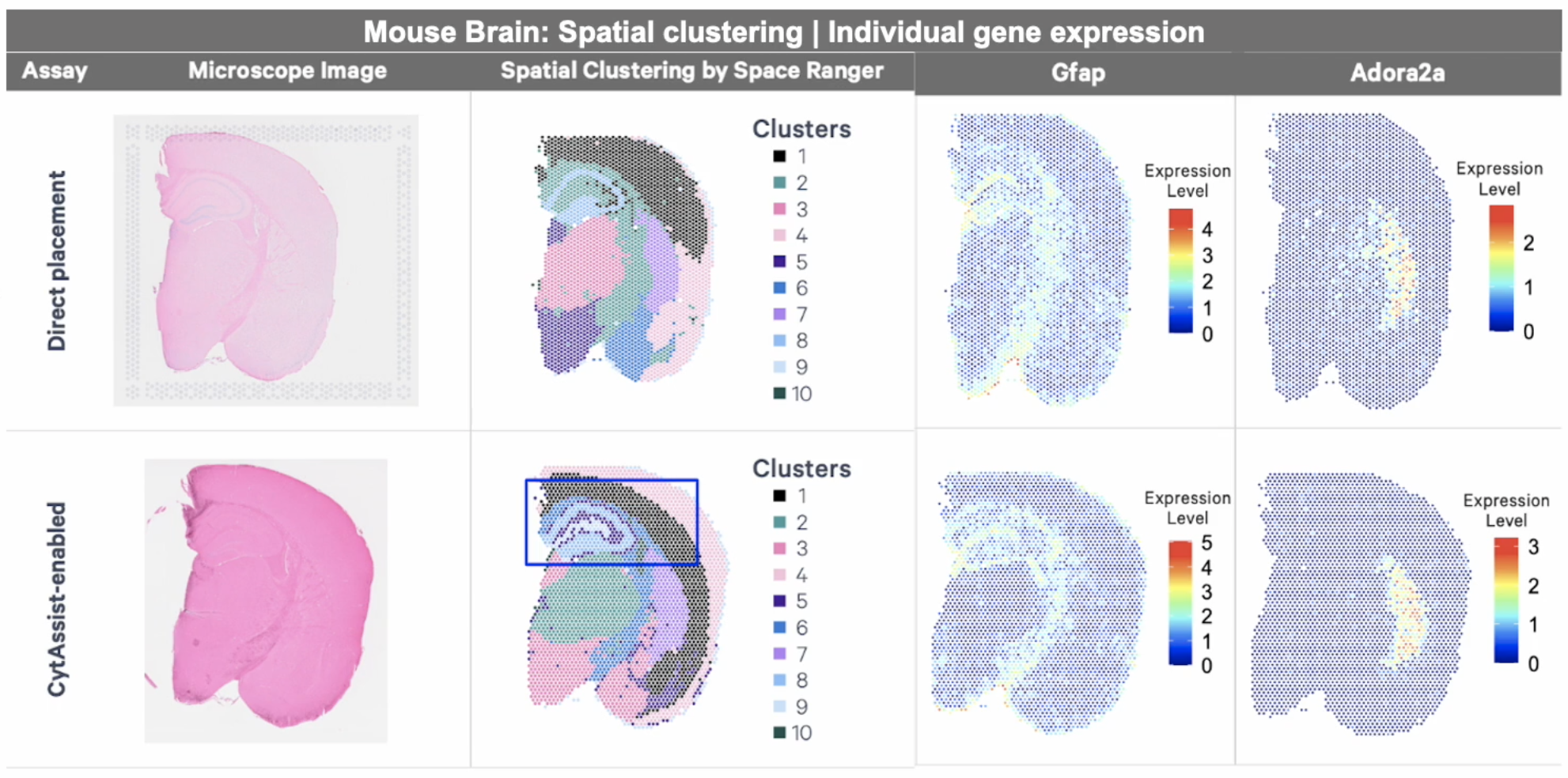
Want to explore the differences between direct placement and CytAssist-enabled assays for FFPE tissue? Review this Technical Note, with a comparison of reagents, workflows, and data between the direct placement and Visium CytAssist-enabled assays.
What samples can I study with Visium CytAssist?
Compatible samples for our CytAssist-enabled v2 Visium Spatial Gene Expression assay match the following conditions:
- Paraformaldehyde (PFA)-fixed frozen, fresh frozen, or formalin-fixed, paraffin-embedded (FFPE) tissue
- Human or mouse
- Tissue sections of varying sizes, with flexibility to analyze an area of interest up to 11 x 11 mm.
(Please note, for the CytAssist-enabled Visium HD assay, compatible samples are human or mouse fresh frozen, fixed frozen, and FFPE tissues and tissue sections of varying sizes with flexibility to analyze an area of interest up to 6.5 x 6.5 mm. See our demonstrated protocols for fresh frozen and fixed frozen Visium HD compatibility.)
With these capabilities, Visium CytAssist offers the most flexible spatial workflow to fit your samples, making a way for analysis of difficult or even traditionally inaccessible sample types. This includes archived, clinical FFPE tissue sections, as well as tissue microarrays.
While FFPE treatment is a popular tissue preservation method, it’s not the only one available to you: fresh frozen and fixed frozen tissues are compatible with v2 Visium CytAssist assays, with minor workflow differences for tissue preparation steps. In house validation experiments have also demonstrated highly sensitive UMI capture among the three sample preparations and strong correlation of genes captured.
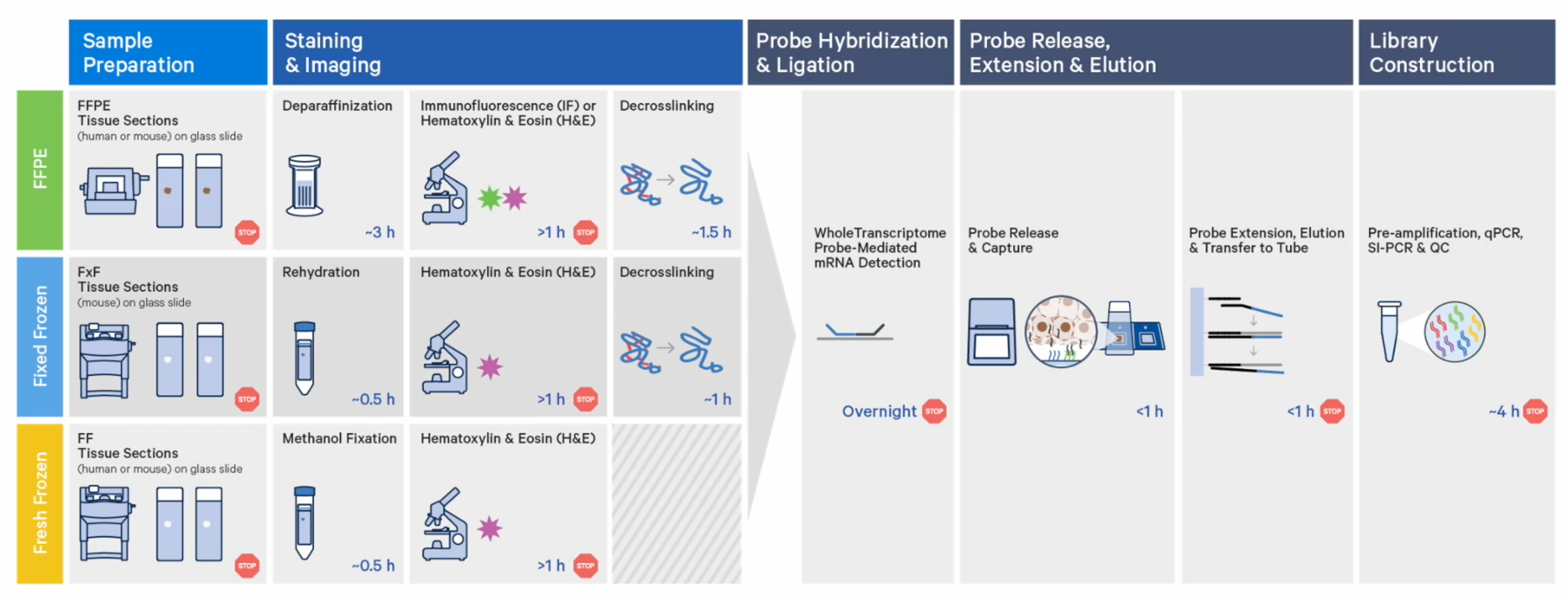
Choosing between these sample preparation options for your spatial experiment really comes down to what tissues you have available to you, and what staining conditions you’d like to employ. (Of note, immunofluorescence staining is only compatible with FFPE tissue at this time.)
Read how researchers at the University of Michigan Advanced Genomics Core are expanding their spatial sample repertoire with Visium CytAssist →
What additional spatial capabilities are enabled on Visium CytAssist?
What would it look like to make the most of precious tissue samples? That was an important question for our teams during development of the Visium CytAssist Gene and Protein Expression assay, which offers protein and whole transcriptome analysis together in a single experiment, from a single tissue section. With the additional protein readout, researchers can better resolve complex cellular microenvironments, such as immune-infiltrated tumor tissues.
Visium CytAssist Gene and Protein Expression uses probe-based ligation chemistry to capture RNA transcripts, with the addition of oligo-tagged antibodies to enable protein co-detection. As part of the product kit, researchers receive a pre-validated, 35-plex antibody immune panel optimized for use on human FFPE tissues. This panel can be combined with additional antibodies against proteins of interest.
During our webinar, there were a number of questions about the antibody panel and spike-in protein detection capabilities:
What is the maximum number of proteins that can be analyzed with Visium CytAssist Gene and Protein Expression?
On top of the 35-plex antibody panel, custom antibodies can be optimized for addition to the assay. The upper limit number of additional targets will vary based on antibody quality and required volumes.
What is the criteria for a good antibody?
Any antibody that works well for immunofluorescence. Once you conjugate the antibodies with oligos, you perform immunofluorescence, and if you’re getting a good signal, then it is very likely that antibody is ‘good’ and is going to work well with the assay.
What, if any, are the steps to optimize the conditions for detecting genes and proteins?
You will not have to optimize anything for gene expression. For proteins, if you’re adding on antibodies, then we have a demonstrated protocol to determine the optimal titer for your antibodies. Once you have that titer, you should be able to pool all those antibodies, along with the immune panel antibodies. It should not require much optimization after you’ve identified the best titer by immunofluorescence.
Will adding protein affect gene expression detection?
No, we have not seen any problems with gene expression once you add proteins. So you will get exactly the same kind of data as if you were only detecting gene expression.
Increased spatial resolution is another approach to maximize the information that you can gain from precious tissue samples. In the webinar, we shared some updates about Visium HD, our upcoming CytAssist-enabled spatial gene expression assay that provides single cell-scale resolution. Stay tuned for more details about Visium HD, coming soon.
What’s an example of the power of Visium CytAssist?
In the first customer publication using Visium CytAssist (1), lead authors Drs. Ryuta Watanabe and Noriyoshi Miura, from Ehime University Hospital in Japan and the Fred Hutchinson Cancer Center, described an experiment to map the spatial gene expression differences between two tumor regions in the same human FFPE prostate cancer sample.
The two regions were molecularly unique cancer subtypes: androgen receptor pathway-positive adenocarcinoma of the prostate (ARPC) and neuroendocrine prostate carcinoma (NEPC). While ARPC is a common form of prostate cancer, NEPC accounts for only 1% of all prostate cancers, making this co-occurring tumor incredibly rare.
With the aid of Visium CytAssist Spatial Gene Expression, the team defined differential gene expression between the tumor subtypes present in the same tissue section, ultimately providing guidance for NEPC diagnosis and potential treatments.
For even more great examples of the power of Visium CytAssist, explore research posters here.
And watch our recent webinar (skip ahead to 24:00 in the recording) to hear how Dr. Lynn van Olst from the Gate Lab at Northwestern University is using Visium CytAsssist Spatial Gene Expression (plus the new Gene and Protein Expression assay) to study the effects of amyloid immunization therapies in FFPE Alzheimer’s patient brain samples taken from a clinical trial in the early 2000s.
References:
- Watanabe R, et al. Spatial gene expression analysis reveals characteristic gene expression patterns of de novo neuroendocrine prostate cancer coexisting with androgen receptor pathway prostate cancer. Int J Mol Sci 24: 8955 (2023).
Editor’s note: This article was updated in February 2024 with relevant content additions and links.
