Three large-scale studies that leveraged single cell to maximize insights from patient samples
Some of biology’s most pressing questions—like how a tumor evolves or why certain therapies work for some patients but not others—can’t be answered with small datasets. More and more researchers are conducting large-scale single cell analyses of millions of cells across hundreds of samples to make breakthrough discoveries.
Of course, with great single cell power comes great data complexity. Mitigating batch effects and ensuring consistent quality across samples aren’t trivial tasks. But our single cell solutions were built to tackle these obstacles, clearing your path to impactful insights.
Our tried-and-tested, instrument-supported workflows provide high reproducibility and proven performance, ensuring consistent data quality across runs and timepoints—no matter how many samples, users, or sites. And with newly developed assay features like advanced multiplexing capabilities, we are continuing to drive down costs, making large-scale single cell studies more accessible than ever.
In this blog, we’ll explore a collection of impactful large-scale studies that leveraged our technology to uncover new insights into health and disease. These publications showcase the transformative potential of scRNA-seq when incorporated into large projects—including cell atlasing, large cohort studies, therapeutic monitoring, and biomarker screening.
Finally, we’ll highlight the latest features and capabilities that make large-scale studies with Chromium a recipe for success, including:

Study 1: Multiomic analysis of large patient cohorts reveals signatures of severe disease
To understand the immune responses driving varied COVID-19 outcomes amongst patients, researchers analyzed 780,000 immune cells collected from the blood samples of 130 infected patients, creating a detailed atlas of how immune cells react—and sometimes overreact—to SARS-CoV-2 infection (1). The groundbreaking study, published in Nature Medicine, used our Universal 5’ Gene Expression Assay to capture both transcriptomic data and immune receptor sequences, ultimately delineating how adaptive immune cells—like T and B cells—coordinate antiviral defenses and contribute to inflammation, which often goes unchecked in severe cases.
By integrating single cell capabilities downstream of conventional immunological tools, the team achieved a level of resolution and scalability that traditional methods, like flow sorting, couldn’t match. These approaches also proved significantly more cost-effective, as conventional workflows would have been far more expensive at this scale.
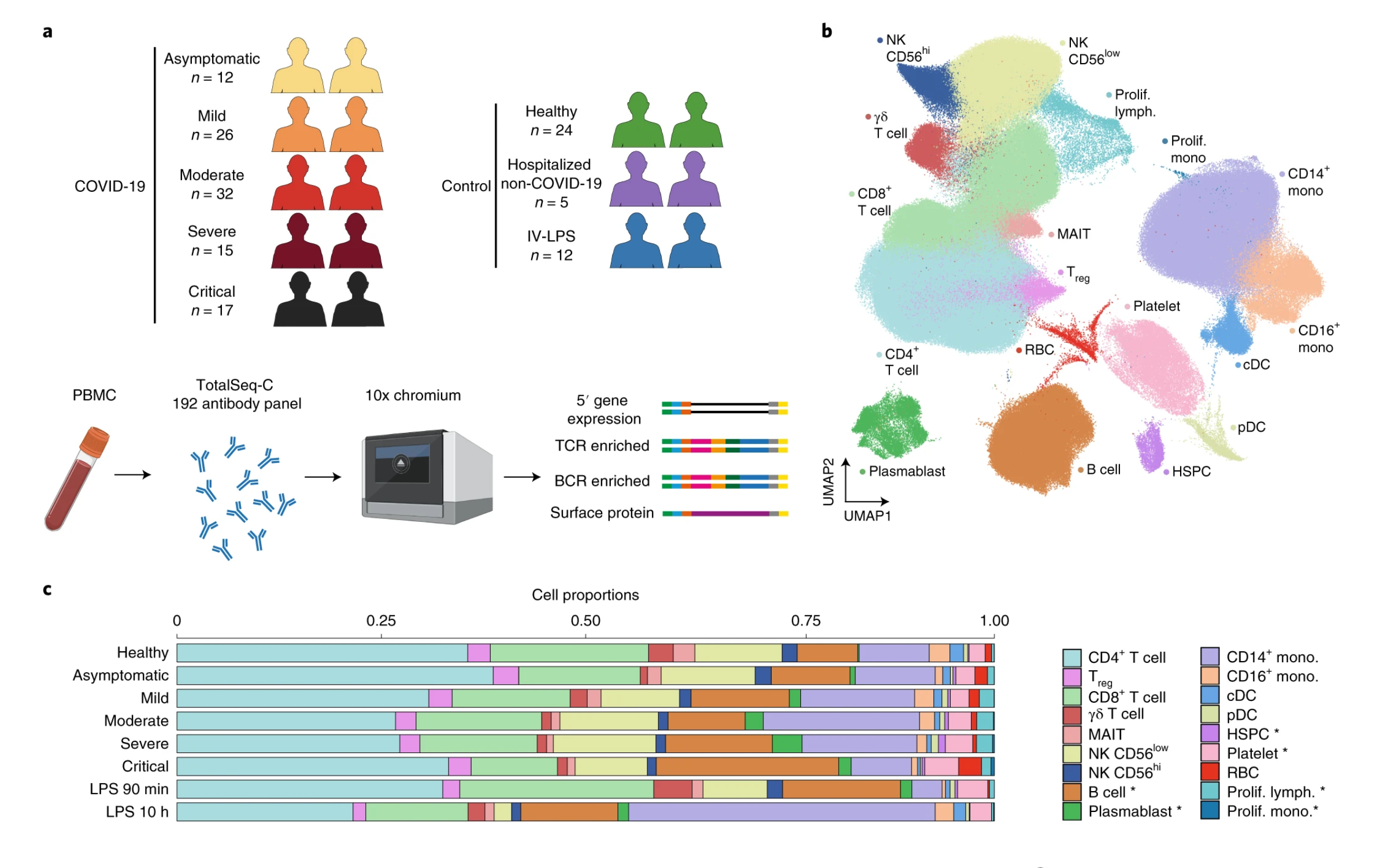
Analyzing nearly a million cells across over 100 patient samples with tried-and-trusted Chromium Single Cell ensured the researchers didn’t overlook any rare cell types and enabled them to conduct meaningful comparisons across diverse patient groups—including varying disease severity and patient demographics. They identified T-cell and B-cell clones associated with mild and severe disease.
The sheer scale of their study, combined with the unmatched sensitivity of our Universal 5’ Gene Expression Assay, revealed a rare immune cell subset of hyperactivated monocytes that were enriched in severe cases and linked to cytokine storms, a potentially life-threatening overreaction of the immune response to viruses. This rare cell subset could be a putative therapeutic target or biomarker for severe cases of COVID-19.
Study 2: Single cell analysis of more than 1 million cells reveals biomarkers of disease severity
Investigators from the University of Pennsylvania used our super sensitive Universal 3’ Gene Expression Assay to pursue a similar question in their study—how does immune dysregulation contribute to disease severity in COVID-19 patients? Ultimately, they sought to make a big impact by identifying potential biomarkers and targets to guide future therapeutic strategies (2).
The researchers also went big with their study, now published in Science, analyzing over a million cells from over 100 hospitalized COVID-19 patients. They opted to analyze a variety of sample types—including PBMCs, nasopharyngeal swabs, and bronchoalveolar lavage fluid.
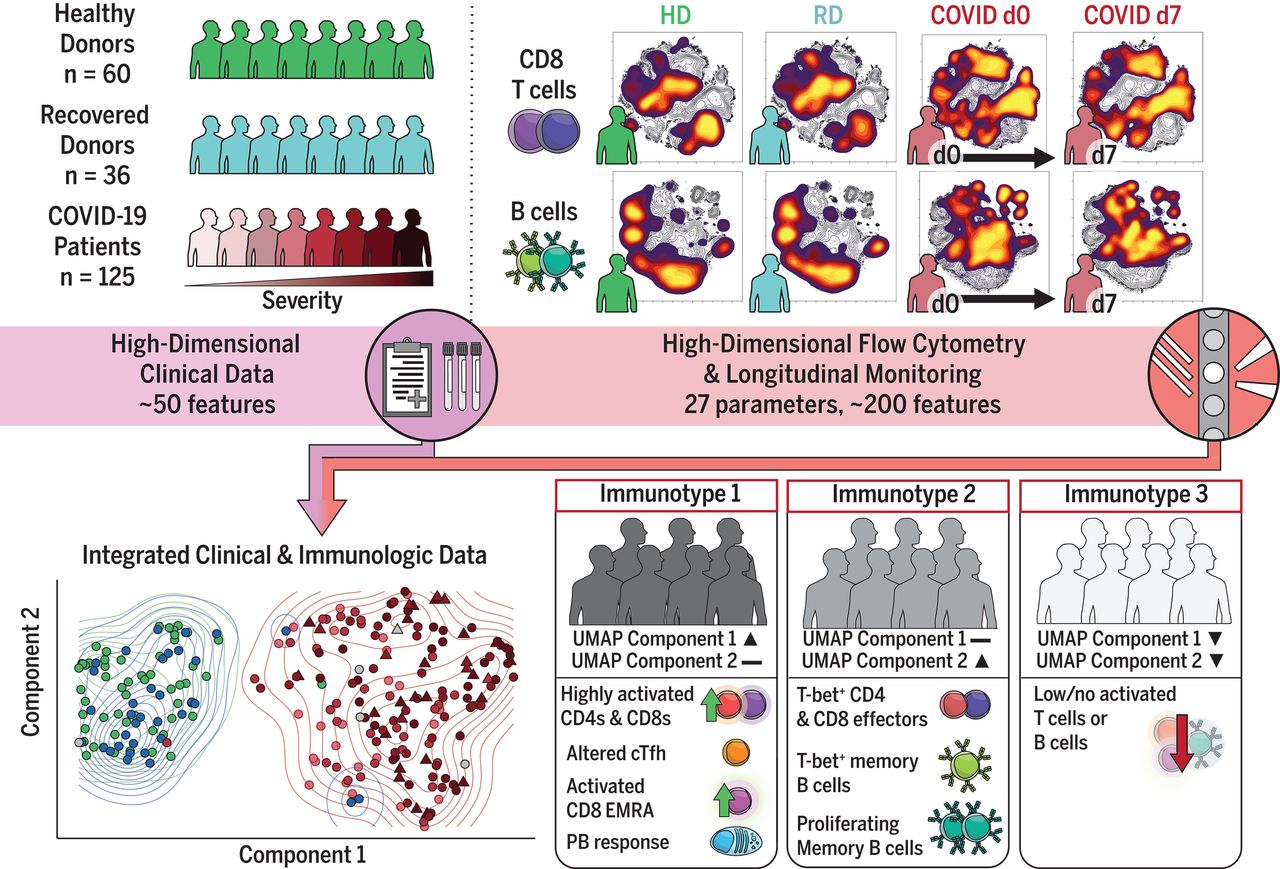
Because our flexible workflows support the broadest range of samples, the researchers could perform high-throughput processing while still providing consistent sensitivity across their hundreds of samples, ensuring robust data quality while minimizing batch effects. This allowed researchers to uncover rare cell populations and confidently connect immune cell behaviors to disease severity.
Ultimately, the authors uncovered different cellular dynamics driving immune responses in patients with mild and severe cases of COVID-19. Their analysis revealed that a population of hyperactivated monocytes contributed to the detrimental cytokine storms often seen in severe cases of COVID-19. In some samples from patients with severe cases, they identified a population of exhausted T cells that impaired immune responses. Conversely, they observed robust T-cell activity in patients with milder outcomes, shedding light on protective immune mechanisms.
The investigators were able to identify therapeutically relevant immune biomarkers of COVID-19 severity and outcomes using a robust data integration strategy. Each patient’s single-cell data was annotated with detailed clinical metadata, including disease severity (mild, moderate, or severe), demographics, treatment regimens, and key biomarkers such as cytokine levels. By associating cellular transcriptional profiles with clinical metadata, they could identify immune cell states and transcriptional changes tied to disease progression and outcomes.
By linking high-dimensional scRNA-seq data with clinical metrics, this study bridged the gap between molecular insights and patient-level outcomes. The identified immune dysregulation patterns driving severe disease provide a roadmap for future studies developing targeted therapies and diagnostic biomarkers.
Study 3: Longitudinal study uncovers the hidden drivers of therapeutic outcomes with scRNA-seq
Researchers from the University of Lausanne and Lausanne University Hospital applied single cell technology to tackle a critical challenge: unraveling immune dynamics longitudinally in cancer patients undergoing adoptive cell therapy (ACT; 3).
The collected samples from 13 patients with metastatic melanoma treated with adoptive cellular therapy with tumor-infiltrating lymphocytes (TIL-ACT) in a phase 1 clinical study (NCT03475134) at key timepoints: at screening (if feasible), at surgery, at minimum 30 days after TIL-ACT, after 4 weeks of nivolumab treatment if applicable (optional), and during progression (optional).
Then, they analyzed the collected samples using the Universal 3’ Gene Expression Assay alongside bulk RNA-seq and spatial proteomics to uncover the mechanisms driving therapeutic success in responders and resistance in nonresponders. Following the patients for over three years provided a comprehensive view of how the tumor microenvironment (TME) evolved with treatment.
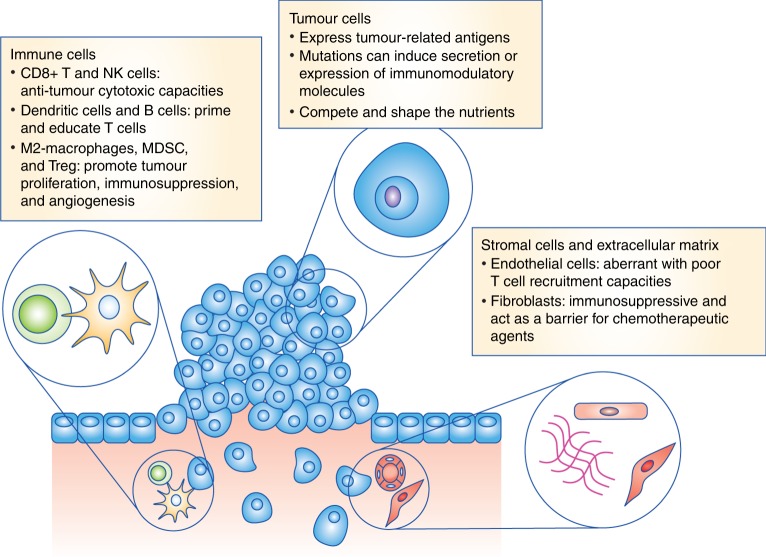
While bulk RNA-seq provided a broad overview of gene expression and spatial proteomics localized immune cell interactions within the TME, the study’s conclusions hinged on the granular insights gained with single cell. Specifically, the researchers discovered that responders possessed preexisting immune niches—composed of tumor-reactive CD8+ T cells and CXCL9+ macrophages—enriched with costimulatory and antigen-presentation signals essential for antitumor immunity. Single cell data revealed transcriptional programs driving these interactions and identified rare, functionally distinct immune subsets, insights that bulk and spatial methods alone could not resolve.
The longitudinal design further demonstrated how Chromium Single Cell solutions excel at tracking immune evolution over time. The Universal 3’ Gene Expression Assay enabled researchers to monitor how treatment reprogrammed the TME in responders, reconstituting antitumor immune networks and reshaping macrophage states to sustain CD8+ T-cell activity.
In contrast, nonresponders lacked these supportive immune interactions, leading to poor therapeutic outcomes. Without single cell resolution, the study could not have linked spatially enriched immune cells to their functional roles or identified key biomarkers, such as CD8+ TIL-macrophage interactions, that predict treatment success.
These findings highlight how single cell technology is indispensable for dissecting complex immune mechanisms in longitudinal studies. By providing the resolution and scalability needed to track immune dynamics across patients and timepoints, the researchers uncovered actionable insights that could improve patient selection and optimize immunotherapy strategies, paving the way for more effective cancer treatments.
Moving large-scale studies forward with single cell innovation
The studies highlighted here are only a few examples of the incredible insights researchers have gained by seamlessly incorporating Chromium Single Cell into their high-throughput studies analyzing samples across patients, timepoints, and institutions.
Researchers are currently collaborating with us to conduct ambitious studies as well, taking advantage of our most advanced single cell technology released in early 2024—GEM-X, which took our proven performance to the next level, as detailed here.
For example, the Garvan Institute of Medical Research will use our Universal 3’ Gene Expression Assay as part of the TenK10K project, which aims to map 50 million human cells across 10,000 individuals to identify unique transcriptional signatures associated with a complex diseases including autoimmune disorders and cancer (5,6).
And we can’t wait to see what more researchers will do with high-quality single cell data from our GEM-X Single Cell Assays, as we expand access with cost-effective solutions, increase input with massive multiplexing, and expand automation support.
Learn more about our new offerings below, and visit our new large projects page to connect with a specialist about how you can elevate the impact of your next ambitious study with single cell analysis.
References:
- Lieberman J, et al. Multiomic analysis of a large cohort of COVID-19 patients reveals signatures of severe disease. Nature Medicine 27: 1340–1355 (2021). doi: 10.1038/s41591-021-01329-2
- Zhang H, et al. Immune dysregulation and therapeutic targets in severe COVID-19: Insights from single-cell analysis. Science 375: 314–321 (2021). doi: 10.1126/science.abc8511
- Barras D, et al. Response to tumor-infiltrating lymphocyte adoptive therapy is associated with preexisting CD8+ T-myeloid cell networks in melanoma. Science Immunology 9: eadg7995 (2024). doi: 10.1126/sciimmunol.adg7995
- Giraldo N, et al. The clinical role of the TME in solid cancer. Br J Cancer 120: 45–53 (2019). doi: 10.1038/s41416-018-0327-z
- 10x Genomics. 10x Genomics Technology Supports TenK10K Project Led by Garvan Institute of Medical Research to Help Transform the Treatment of Complex Diseases. News Release. August 2024. Available at: https://investors.10xgenomics.com/news/news-details/2024/10x-Genomics-Technology-Supports-TenK10K-Project-Led-by-Garvan-Institute-of-Medical-Research-to-Help-Transform-the-Treatment-of-Complex-Diseases/default.aspx
- D’Urso M, et al. Single-cell eQTL mapping identifies cell type–specific genetic control of autoimmune disease. Science 376: 555–562 (2024). doi: 10.1126/science.abf3041
About the author:

