Space Ranger analyzes sequencing data produced from Visium Spatial Gene Expression for fresh frozen (FF) tissue samples. The workflow diagram shows all the required inputs and the corresponding parameters for FF tissue samples:
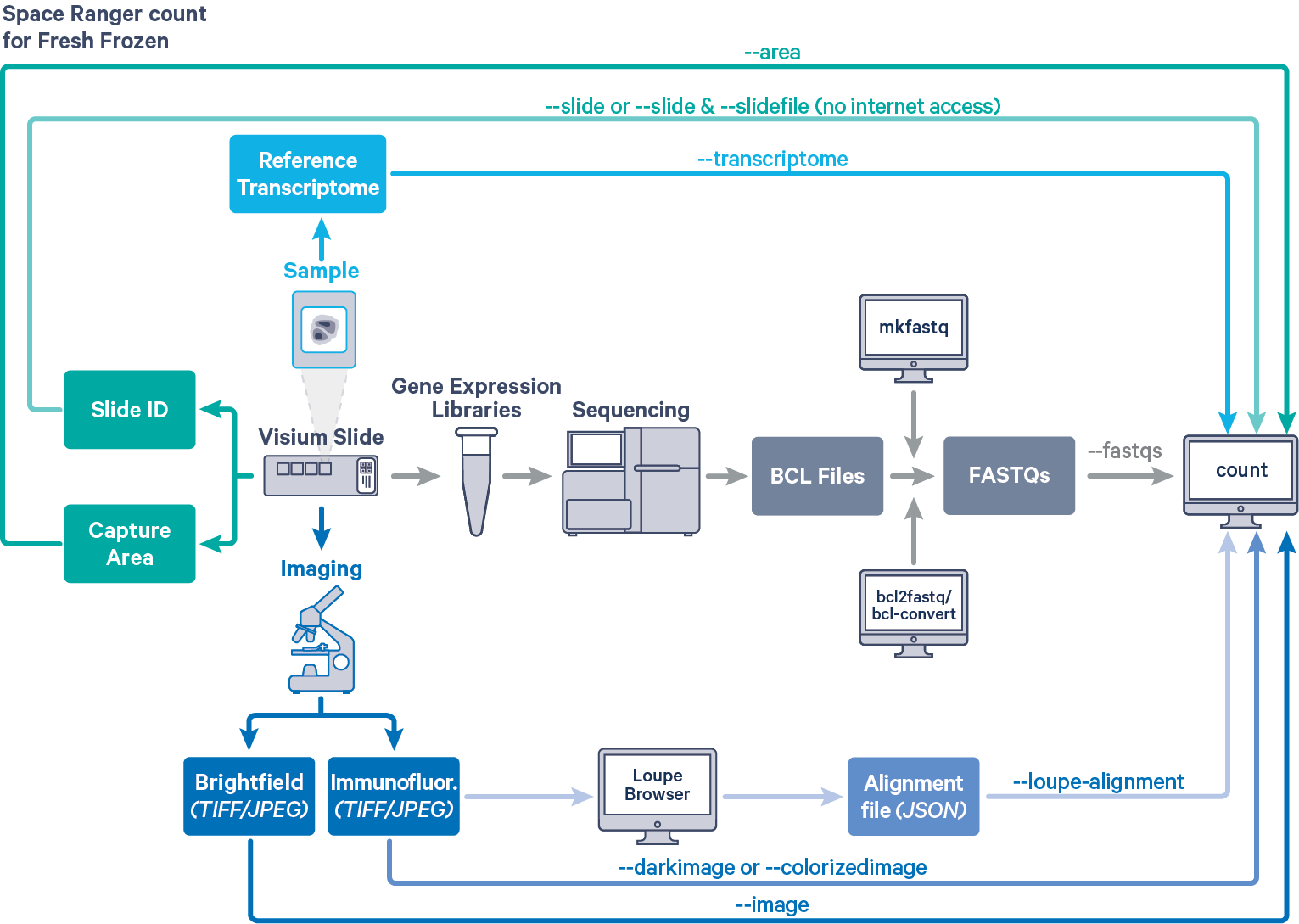
The spaceranger count pipeline is run on every Capture Area in the Visium slide. The required inputs are:
- The FASTQ files for a given capture area (
--fastqs)- If starting with Illumina BCL output folder, follow the instructions on running spaceranger mkfastq to generate FASTQ files
- For help on which arguments to use to target a particular set of FASTQs, consult Specifying Input FASTQ Files for 10x Pipelines
- The corresponding capture area image in either
"TIFF,"QPTIFFor"JPEGformat--imagefor brightfield image--darkimagefor dark background fluorescence image--colorizedimagefor composite colored fluorescence image
- The slide layout GPR file specified by
--slide&--areaifspacerangerhas access to internet--slidefile,--slide&--areaifspacerangerhas no access to internet. The slide layout file is directly downloaded--unknown-slideif visium slide details are unknown
- The reference transcriptome for the species from which the sample was obtained (
--transcriptome)
For a list of accepted arguments, see the command line arguments](/support/software/space-ranger/3.0/analysis/running-pipelines/command-line-arguments), or run spaceranger count --help. The following code examples assume compute platforms are connected to the internet wherespaceranger uses the value of the --slide argument to automatically download the slide layout file in gpr format. In absence of internet connectivity to the compute platform, the user can download the slide specific layout file in gpr format and provide it to spaceranger using the --slidefile argument. Refer to the Running spaceranger count (FF) tutorial for detailed walkthrough.
Select the section corresponding to the imaging workflow.
To generate spatial feature counts for a single library from fresh frozen (FF) sample using automatic fiducial alignment and tissue detection on a brightfield image input, run spaceranger count with the following arguments.
cd /home/jdoe/runs
spaceranger count --id=sample345 \ #Output directory
--transcriptome=/home/jdoe/refdata/GRCh38-2020-A \ #Path to Reference
--fastqs=/home/jdoe/runs/HAWT7ADXX/outs/fastq_path \ #Path to FASTQs
--sample=mysample \ #Sample name from FASTQ filename
--image=/home/jdoe/runs/images/sample345.tiff \ #Path to brightfield image
--slide=V19J01-123 \ #Slide ID
--area=A1 \ #Capture area
--localcores=8 \ #Allowed cores in localmode
--localmem=64 #Allowed memory (GB) in localmode
Following a set of preflight checks to validate the input arguments, spaceranger count will begin to run.
To generate spatial feature counts for a single library fresh frozen (FF) sample using a fiducial alignment and tissue assignment JSON file generated in Loupe Browser on a brightfield image input, run spaceranger count with the following arguments. The highlighted code indicates inputs that do not change.
1cd /home/jdoe/runs 2spaceranger count --id=sample345 \ #Output directory 3 --transcriptome=/home/jdoe/refdata/GRCh38-2020-A \ #Path to Reference 4 --fastqs=/home/jdoe/runs/HAWT7ADXX/outs/fastq_path \ #Path to FASTQs 5 --sample=mysample \ #Sample name from FASTQ filename 6 --image=/home/jdoe/runs/images/sample345.tiff \ #Path to brightfield image 7 --slide=V19J01-123 \ #Slide ID 8 --area=A1 \ #Capture area 9 --loupe-alignment=sample345.json \ #Manual fiducial alignment file 10 --localcores=8 \ #Allowed cores in localmode 11 --localmem=64 #Allowed memory (GB) in localmode
The arguments to specify the fluorescence image can be either --darkimage or --colorizedimage depending on the image format. Details about the different supported fluorescence image formats are described in Image Recommendations.
Following a set of preflight checks to validate input arguments, spaceranger count pipeline stages will begin to run.
A successful spaceranger count run concludes with a message similar to this:
- Run summary HTML: /home/jdoe/runs/sample345/outs/web_summary.html
- Outputs of spatial pipeline:
aligned_fiducials: /home/jdoe/runs/sample345/outs/spatial/aligned_fiducials.jpg
detected_tissue_image: /home/jdoe/runs/sample345/outs/spatial/detected_tissue_image.jpg
scalefactors_json: /home/jdoe/runs/sample345/outs/spatial/scalefactors_json.json
tissue_hires_image: /home/jdoe/runs/sample345/outs/spatial/tissue_hires_image.png
tissue_lowres_image: /home/jdoe/runs/sample345/outs/spatial/tissue_lowres_image.png
cytassist_image: null
aligned_tissue_image: null
tissue_positions: /home/jdoe/runs/sample345/outs/spatial/tissue_positions.csv
spatial_enrichment: /home/jdoe/runs/sample345/outs/spatial/spatial_enrichment.csv
barcode_fluorescence_intensity: null
- Run summary CSV: /home/jdoe/runs/sample345/outs/metrics_summary.csv
- Correlation values between isotypes and Antibody features: null
- BAM: /home/jdoe/runs/sample345/outs/possorted_genome_bam.bam
- BAM BAI index: /home/jdoe/runs/sample345/outs/possorted_genome_bam.bam.bai
- BAM CSI index: null
- Filtered feature-barcode matrices MEX: /home/jdoe/runs/sample345/outs/filtered_feature_bc_matrix
- Filtered feature-barcode matrices HDF5: /home/jdoe/runs/sample345/outs/filtered_feature_bc_matrix.h5
- Unfiltered feature-barcode matrices MEX: /home/jdoe/runs/sample345/outs/raw_feature_bc_matrix
- Unfiltered feature-barcode matrices HDF5: /home/jdoe/runs/sample345/outs/raw_feature_bc_matrix.h5
- Secondary analysis output CSV: /home/jdoe/runs/sample345/outs/analysis
- Per-molecule read information: /home/jdoe/runs/sample345/outs/molecule_info.h5
- Loupe Browser file: /home/jdoe/runs/sample345/outs/cloupe.cloupe
- Feature Reference: null
- Target Panel file: null
- Probe Set file: null
Pipestance completed successfully!
The output of the pipeline is contained in a folder named with the sample ID specified using --id (e.g. sample345). If this folder already exists, in a rerun with the original parameters, spaceranger will assume it is an existing pipestance and attempt to resume running it. The subfolder named outs contains the main pipeline output files.
Once spaceranger count has successfully completed, you can further explore the results by
- Reviewing the web_summary.html
- Opening the
.cloupefile in Loupe Browser - Referring to the Understanding Outputs page