Space Ranger relies on image processing algorithms to solve two key problems with respect to the slide image: deciding where tissue has been placed and aligning the printed fiducial spot pattern. Tissue detection is needed to identify which capture spots, and therefore which barcodes, will be used for analysis. Fiducial alignment is needed so Space Ranger can know where in the image an individual barcoded spot resides, since each user may set a slightly different field of view when imaging the Visium capture area.
These problems are made difficult by the fact that the image scale and size are unknown, an unknown subset of the fiducial frame may be covered by tissue or debris, each tissue has its own unique appearance, and the image exposure may vary from run to run.
If either of the procedures described here fail to perform adequately, the user can use the manual alignment and tissue selection wizard in Loupe Browser. In most cases, the pipeline is not able to indicate that a failure has occurred and the user is encouraged to check the quality-control image in the Summary tab of the Web Summary that Space Ranger outputs. Note that the user supplied full resolution original image is used for downstream visualization in Loupe Browser.
Finally, Space Ranger v2.0 onwards the automatic image analysis described here, will have --reorient-images=true by default which will automatically rotate and mirror the image such that the "hourglass" fiducial is in the upper-left (see details in Fiducial alignment):
| Properly Oriented Slide Image | Corner with Hourglass Fiducial |
|---|---|
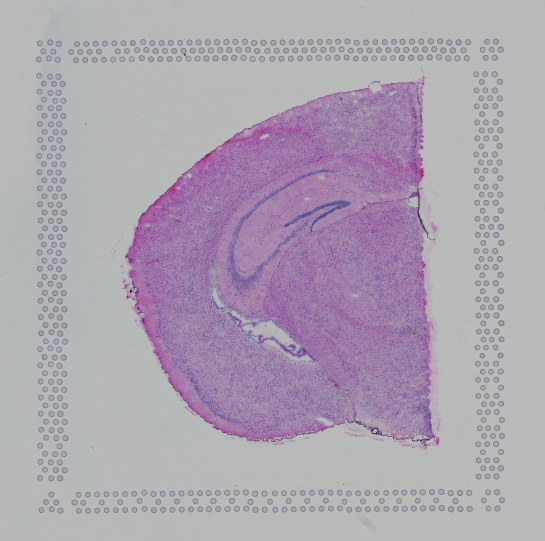 | 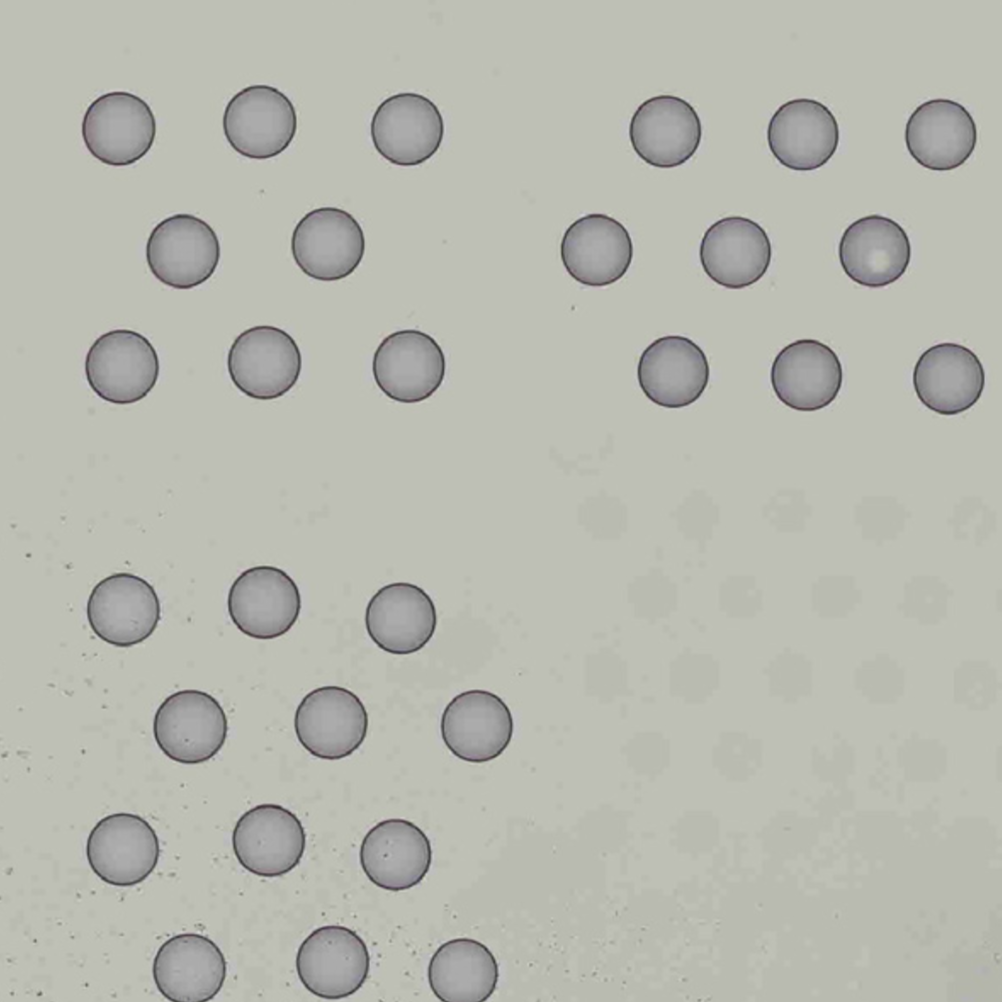 |
Visium contains a system for identifying the slide-specific pattern of invisible capture spots printed on each slide and how these relate to the visible fiducial spots that form a frame around each capture area. The fiducial frame has unique corners and sides that the software attempts to identify.
This process first extracts features that "look" like fiducial spots and then attempts to align these candidate fiducial spots to the known fiducial spot pattern. The spots extracted from the image will necessarily contain some misses, for instance in places where the fiducial spots were covered by tissue, and some false positives, such as where debris on the slide or tissue features may look like fiducial spots.
After extraction of putative fiducial spots from the image, this pattern is aligned to the known fiducial spot pattern in a manner that is robust to a reasonable number of false positives and false negatives. The output of this process is a coordinate transform that relates the Visium barcoded spot pattern to the user's tissue image.
From Space Ranger v2.0 onwards the pipeline is run with the flag --reorient-images=true by default which removes the constraint of the hourglass fiducial marker needing to be in the upper-left corner. This is accomplished by running the alignment algorithm against each of the eight possible fiducial frame transformations (all rotations by 90 degrees plus mirroring) and choosing among those the
alignment with the best fit.
Each area in the standard Visium slide contains a grid of 4,992 capture spots and 14,336 capture spots in the XL Visium slide, all of which are populated with spatially barcoded probes for capturing poly-adenylated mRNA. In practice, only a fraction of these spots are covered by tissue. In order to restrict Space Ranger's analysis to only those spots where tissue was placed, Space Ranger uses an algorithm to identify tissue in the input brightfield image.
Using a grayscale, downsampled version of the input image while maintaining the original aspect ratio, multiple estimates of tissue section placement are calculated and compared. These estimates are used to train a statistical classifier to label each pixel within the capture area as either tissue or background.
The quality control image below shows the result of this process. Individual image pixels determined to be tissue are colored red. The locations of the invisible capture spots are drawn as gray circles when over background.
| Quality Control Image |
|---|
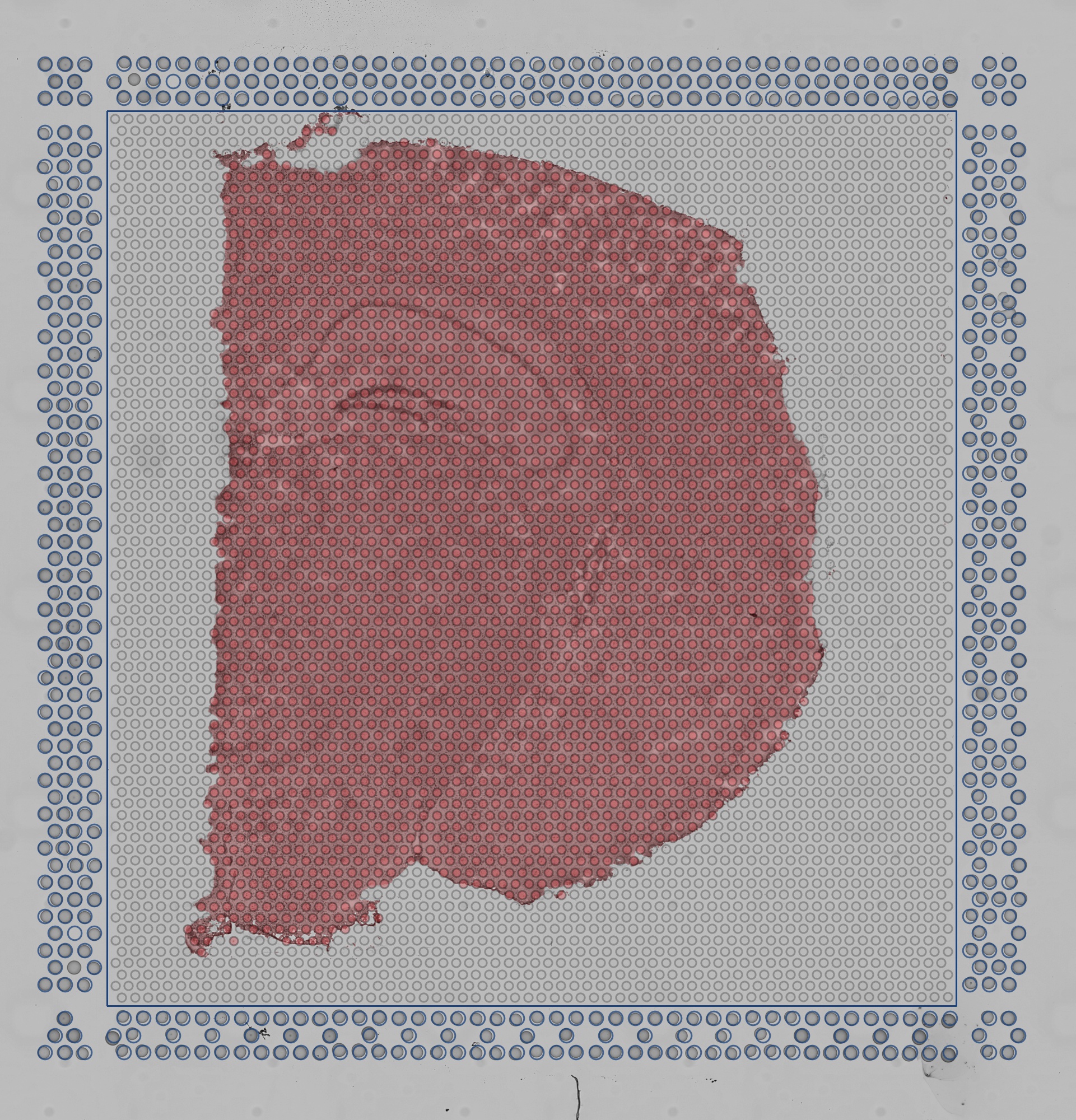 |
In order to achieve optimal results, the algorithm expects an image with a smooth, bright background and darker tissue with a complex structure. If the area drawn in red does not coincide with the tissue, you can perform manual alignment and tissue selection in Loupe Browser before running Space Ranger. See the Imaging Technical Note for information about how to optimize imaging conditions for automated alignment and tissue detection.
Support for CytAssist enabled Gene Expression analysis was introduced in Space Ranger v2.0. This workflow includes the use of CytAssist image which has the fiducial frame and optionally, a microscope image (brightfield or fluorescence image types) of the same tissue section on the standard glass slide. When the two image inputs are provided, the images need to be registered for downstream visualization.
The targeted transformation of the image registration process is the 2D similarity transformation, which includes homogeneous scaling, translation, rotation, and mirroring. To enable automated tissue image registration between the microscope image and CytAssist image, the algorithm first estimates the proper scaling and translation so the two tissue sections in the images can be roughly overlaid together. Since the correct rotation and mirroring are hard to estimate, the pipeline addresses this by testing eight possible transformations (all rotations by 90 degrees plus mirroring). The estimated scaling, translation, and rotation are used as the initialization to start the registration which is an optimization process. The optimization relies on the mutual information metric from the open source software ITK. When optimization of all different initializations are complete, the pipeline chooses the best combination based on the smallest metric (entropy) of Mattes mutual information to output as the final registration results.
The resulting quality control image below captures the results of the image registration represented as a checkerboard pattern in which the alternating squares represent the two different image inputs.
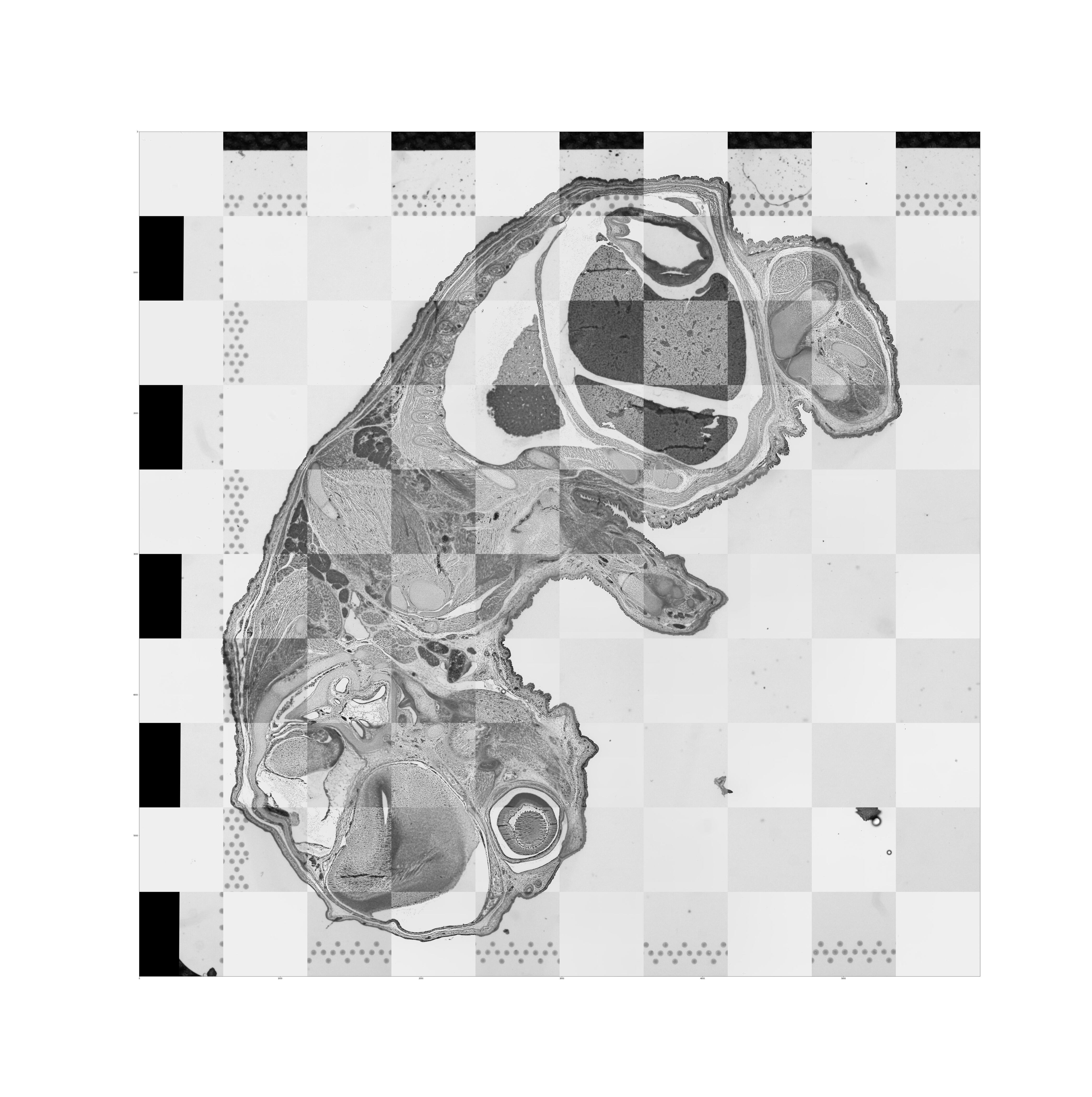
From Space Ranger 2.0 onwards fluorescence intensity in the image at every barcoded-spot location is quantified and is captured in the barcode_fluorescence_intensity.csv file. Each page of an input grayscale image is downsampled to the same dimensions as the tissue_hires_image.png image that can be found in the outs/spatial directory. From there the pixels are assigned to each barcoded-spot for each page. The location and size of each spot is defined in tissue_positions.csv and scalefactors_json.json. Space Ranger then calculates the mean and standard deviation of the intensities from pixels that are entirely within the area of the known barcoded-spot location for each page.