The cellranger mkfastq pipeline is deprecated and will be removed in a future release. Please use Illumina’s BCL Convert to generate Cell Ranger-compatible FASTQ files. For detailed guidance, refer to the Generating FASTQs page.
The Cell Ranger workflow starts by demultiplexing the Illumina sequencer's base call files (BCLs) for each flow cell directory into FASTQ files. 10x Genomics has developed cellranger mkfastq, a pipeline that wraps Illumina's bcl2fastq and provides a number of convenient features in addition to the features of bcl2fastq: - Translates 10x Genomics sample index names into the corresponding oligonucleotides in the sample index - Supports a simplified CSV sample sheet format to handle 10x Genomics use cases - Supports most bcl2fastq arguments, such as --use-bases-mask
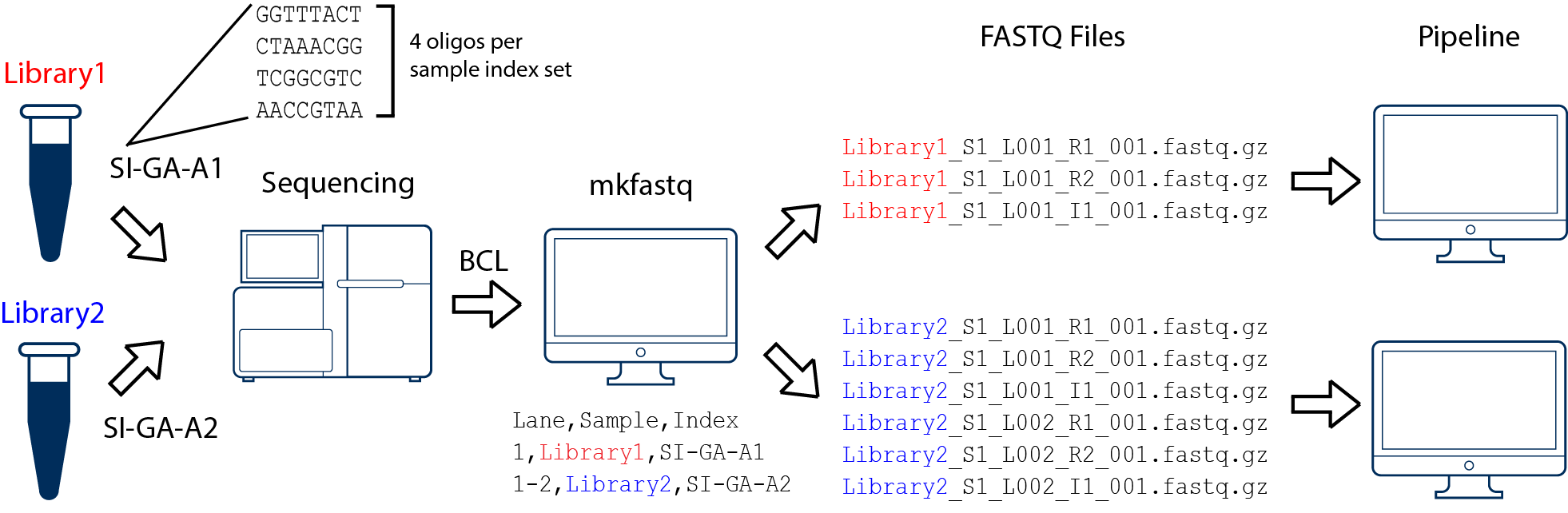
Cell Ranger's mkfastq supports single-indexed and dual-indexed flow cells. Single and dual-indexed samples should be processed in separate instances of the cellranger mkfastq pipeline. The pipeline will select the appropriate mode depending on the sample indexes used, and enable index-hopping filtering automatically for dual-indexed flow cells. For example, with the Dual Index Kit TT Set A, well A1 can be specified in the sample sheet as "SI-TT-A1", and cellranger mkfastq will recognize the i7 and i5 indices as GTAACATGCG and AGTGTTACCT, respectively. Similarly for Single Index Kit T Set A, well A1 can be specified in the sample sheet as "SI-GA-A1", and cellranger mkfastq will recognize the four i7 indexes (GGTTTACT, CTAAACGG, TCGGCGTC, and AACCGTAA) and merge the resulting FASTQ files.
In this example, we have two 10x Genomics libraries (each processed through a separate Chromium chip channel) that are multiplexed on a single flow cell. Note that after running cellranger mkfastq, we run a separate instance of the cellranger pipeline on each library.
In this example, we have one 10x Genomics library sequenced on two flow cells. Note that after running cellranger mkfastq, we run a single instance of the cellranger pipeline on all the FASTQ files generated.

The cellranger mkfastq pipeline accepts additional options beyond those shown in the table below because it is a wrapper around bcl2fastq. Consult the User Guide for Illumina's bcl2fastq for more information.
| Parameter | Function |
|---|---|
--run | Required. The path of Illumina BCL run folder. |
--id | Optional. Defaults to the name of the flow cell referred to by --run. Name of the folder created by mkfastq. This option does not accept a full directory path. If used, navigate to the specified directory first and then run mkfastq. |
--samplesheet | Optional. Path to an Illumina Experiment Manager-compatible sample sheet which contains 10x Genomics sample index names (e.g., SI-GA-A1 or SI-TT-A12) in the sample index column. All other information, such as sample names and lanes, should be in the sample sheet. |
--sample-sheet | Optional. Equivalent to --samplesheet above. |
--csv | Optional. Path to a simple CSV with lane, sample, and index columns, which describe the way to demultiplex the flow cell. The index column should contain a 10x Genomics sample dual-index name (e.g., SI-TT-A12). This is an alternative to the Illumina IEM sample sheet, and will be ignored if --samplesheet is specified. |
--simple-csv | Optional. Equivalent to --csv above. |
--filter-dual-index | Optional. Only demultiplex samples identified by i7/i5 dual-indices (e.g., SI-TT-A6), ignoring single-index samples. Single-index samples will not be demultiplexed. Also notice that Cell Ranger will run single-index data, but it is not supported. |
--filter-single-index | Optional. Only demultiplex samples identified by an i7-only sample index, ignoring dual-indexed samples. Dual-indexed samples will not be demultiplexed. |
--lanes | bcl2fastq option. Comma-delimited series of lanes to demultiplex (e.g. 1,3). Use this if you have a sample sheet for an entire flow cell but only want to generate a few lanes for further 10x Genomics analysis. |
--use-bases-mask | bcl2fastq option. Same meaning as for bcl2fastq. Use to clip extra bases off a read if you ran extra cycles for QC. |
--delete-undetermined | bcl2fastq option. Delete the Undetermined FASTQs generated by bcl2fastq. Useful if you are demultiplexing a small number of samples from a large flow cell. |
--barcode-mismatches | bcl2fastq option. Same meaning as for bcl2fastq. Use this option to change the number of allowed mismatches per index adapter (0, 1, 2). Default: 1. |
--output-dir | bcl2fastq option. Generate FASTQ output in a path of your own choosing, instead of flowcell_id/outs/fastq_path. |
--project | bcl2fastq option. Custom project name, to override the sample sheet or to use in conjunction with the --csv argument. |
--jobmode | Martian option. Job manager to use. Valid options: local (default), sge, lsf, or a .template file. |
--localcores | Martian option. Set max cores the pipeline may request at one time. Only applies when --jobmode=local. |
--localmem | Martian option. |
The Cell Ranger mkfastq pipeline recognizes two file formats for describing samples: a simple, three-column CSV format, or the Illumina Experiment Manager (IEM) sample sheet format used by bcl2fastq. There is an example below for running mkfastq with each format.
The example (tiny-bcl) dataset is solely designed to demo the cellranger mkfastq pipeline. It cannot be used to run downstream pipelines (e.g. cellranger count).
To follow along:
- Download the tiny-bcl tar file
- Decompress the tiny-bcl tar file in your working directory. This will create a new
tiny-bclsubdirectory - Download the simple CSV layout file
- Download the Illumina Experiment Manager sample sheet
A simple CSV sample sheet is recommended for most sequencing experiments. The simple CSV format has only three columns (Lane, Sample, Index), and is thus less prone to formatting errors. You can see an example of this in cellranger-tiny-bcl-simple-1.2.0.csv.
Lane,Sample,Index
1,test_sample,SI-TT-D9
If you have multiple library types (e.g., Gene Expression, Feature Barcode, and Cell Multiplexing) that all have the same type of indexing (e.g., dual-indexing), the samples can be demultiplexed together and the CSV could be formatted as follows (the Sample is named by library type here for demonstration only):
Lane,Sample,Index
1,GEX_sample,SI-TT-D9
1,FB_sample,SI-NT-A1
1,CMO_sample,SI-NN-A1
Here are the options for each column:
| header | something |
|---|---|
| Lane | Which lane(s) of the flow cell to process. Can be either a single lane, a range (e.g., 2-4) or "*" for all lanes in the flow cell. |
| Sample | The name of the sample. This name is the prefix to all the generated FASTQs, and corresponds to the --sample argument in all downstream 10x Genomics pipelines. Sample names must conform to the Illumina bcl2fastq naming requirements. Only letters, numbers, underscores, and hyphens are allowed; no other symbols, including dots ("."), are allowed. |
| Index | The 10x Genomics sample index that was used in library construction, e.g., SI-TT-D9 or SI-GA-A1 |
To run mkfastq with a simple layout CSV, use the --csv argument.
Here's how to run mkfastq on the tiny-bcl sequencing run with the simple layout (customize the code with the path to tiny_bcl on your system):
cellranger mkfastq --id=tiny-bcl \
--run=/path/to/tiny_bcl \
--csv=cellranger-tiny-bcl-simple-1.2.0.csv
cellranger mkfastq
Copyright (c) 2019 10x Genomics, Inc. All rights reserved.
-------------------------------------------------------------------------------
Martian Runtime - 7.1.0-v4.0.8
Running preflight checks (please wait)...
2019-11-14 16:33:54 [runtime] (ready) ID.tiny-bcl.MAKE_FASTQS_CS.MAKE_FASTQS.PREPARE_SAMPLESHEET
2019-11-14 16:33:57 [runtime] (split_complete) ID.tiny-bcl.MAKE_FASTQS_CS.MAKE_FASTQS.PREPARE_SAMPLESHEET
2019-11-14 16:33:57 [runtime] (run:local) ID.tiny-bcl.MAKE_FASTQS_CS.MAKE_FASTQS.PREPARE_SAMPLESHEET.fork0.chnk0.main
2019-11-14 16:34:00 [runtime] (chunks_complete) ID.tiny-bcl.MAKE_FASTQS_CS.MAKE_FASTQS.PREPARE_SAMPLESHEET
...
The cellranger mkfastq pipeline can also be run with a sample sheet in the Illumina Experiment Manager (IEM) format (example: cellranger-tiny-bcl-samplesheet-1.2.0.csv. An IEM sample sheet has several fields specific to running on Illumina platforms, including a [Data] section where sample and index information is specified. mkfastq supports listing either index set names or the oligo sequences.
Dual-indexing example
Version 1: "SI-TT-D9" refers to a 10x Genomics dual-index sample index, so mkfastq auto-detects that this is a dual-index sample. In this example, only reads from lane 1 will be used. To demultiplex the given sample index across all lanes, omit the Lane column entirely.
[Data]
Lane,Sample_ID,index
1,test_sample,SI-TT-D9
Version 2: The index sequences for "SI-TT-D9" are specified in the two index and index2 columns.
[Data]
Lane,Sample_ID,index,index2
1,test_sample,TGGTCCCAAG,ACGCCAGAGG
Single-indexing example
Here, "SI-GA-A3" refers to a 10x Genomics single index sample index consisting of a set of four oligo sequences.
[Data]
Lane,Sample_ID,index
1,Sample1,SI-GA-A3
Sample names must conform to the Illumina bcl2fastq naming requirements. Specifically, only letters, numbers, underscores, and hyphens are allowed. No other symbols, including dots ("."), are allowed.
Also note that while an authentic IEM sample sheet will contain other sections above the [Data] section, these are optional for demultiplexing. To avoid data loss from trimming, we do not recommend including adapter sequences in the [Settings] section of the sample sheet (see this article for details). For demultiplexing an existing run with cellranger mkfastq, only the [Data] section is required.
Next, run the cellranger mkfastq pipeline, using the --samplesheet argument (customize the path to tiny_bcl on your system):
cellranger mkfastq --id=tiny-bcl \
--run=/path/to/tiny_bcl \
--samplesheet=cellranger-tiny-bcl-samplesheet-1.2.0.csv
cellranger mkfastq
Copyright (c) 2019 10x Genomics, Inc. All rights reserved.
-------------------------------------------------------------------------------
Martian Runtime - 7.1.0-v4.0.8
Running preflight checks (please wait)...
2019-11-14 16:35:49 [runtime] (ready) ID.tiny-bcl.MAKE_FASTQS_CS.MAKE_FASTQS.PREPARE_SAMPLESHEET
2019-11-14 16:35:52 [runtime] (split_complete) ID.tiny-bcl.MAKE_FASTQS_CS.MAKE_FASTQS.PREPARE_SAMPLESHEET
2019-11-14 16:35:52 [runtime] (run:local) ID.tiny-bcl.MAKE_FASTQS_CS.MAKE_FASTQS.PREPARE_SAMPLESHEET.fork0.chnk0.main
2019-11-14 16:35:58 [runtime] (chunks_complete) ID.tiny-bcl.MAKE_FASTQS_CS.MAKE_FASTQS.PREPARE_SAMPLESHEET
...
If you encounter any preflight errors, refer to the Troubleshooting page.
Once the cellranger mkfastq pipeline has successfully completed, the output can be found in a new folder named with the value you provided to cellranger mkfastq in the --id option (if not specified, defaults to the name of the flow cell):
ls -l
drwxr-xr-x 4 jdoe jdoe 4096 Nov 14 12:05 tiny-bcl
The key output files can be found in outs/fastq_path, and are organized in the same manner as a conventional cellranger bcl2fastq run:
ls -l tiny-bcl/outs/fastq_path/
drwxr-xr-x 3 jdoe jdoe 3 Nov 14 12:26 Reports
drwxr-xr-x 2 jdoe jdoe 8 Nov 14 12:26 Stats
drwxr-xr-x 3 jdoe jdoe 3 Nov 14 12:26 tiny-bcl
-rw-r--r-- 1 jdoe jdoe 20615106 Nov 14 12:26 Undetermined_S0_L001_I1_001.fastq.gz
-rw-r--r-- 1 jdoe jdoe 20615106 Nov 14 12:26 Undetermined_S0_L001_I2_001.fastq.gz
-rw-r--r-- 1 jdoe jdoe 51499694 Nov 14 12:26 Undetermined_S0_L001_R1_001.fastq.gz
-rw-r--r-- 1 jdoe jdoe 152692701 Nov 14 12:26 Undetermined_S0_L001_R2_001.fastq.gz
tree tiny-bcl/outs/fastq_path/tiny_bcl/
tiny-bcl/outs/fastq_path/tiny_bcl/
Sample1
Sample1_S1_L001_I1_001.fastq.gz
Sample1_S1_L001_I2_001.fastq.gz
Sample1_S1_L001_R1_001.fastq.gz
Sample1_S1_L001_R2_001.fastq.gz
This example was produced with a sample sheet that included tiny-bcl as the Sample_Project, so the directory containing the sample folders is called tiny-bcl. If a Sample_Project was not specified, or if a simple layout CSV file was used (which does not have a Sample_Project column), the directory containing the sample folders would be named according to the flow cell ID instead.
If you want to remove the Undetermined FASTQs from the output to save space, you can run mkfastq with the --delete-undetermined flag. To see all cellranger mkfastq options, run cellranger mkfastq --help.
If you encounter a crash while running cellranger mkfastq, upload the tarball (with the extension .mri.tgz) in your output directory. Customize the code with your email:
cellranger upload your@email.edu jobid.mri.tgz
Where jobid is what you input into the --id option of mkfastq (if not specified, defaults to the ID of the flow cell). This tarball contains numerous diagnostic logs that 10x Genomics support can use for debugging.
You will receive an automated email from 10x Genomics. If not, email support@10xgenomics.com. For the fastest service, respond with the following:
- The exact
cellrangercommand you used - The sample sheet that you used
- The
RunInfo.xmlandrunParameters.xmlfiles from your BCL directory - The kind of libraries you are demultiplexing (including chemistry)