Spatially resolved transcriptomics: An introductory overview of spatial gene expression profiling methods
Spatially resolved transcriptomics—named the 2020 Method of the Year by Nature Methods (1)—is a rising star. Still, if you are new to the field, you may ask: why is profiling the spatial location of biological components essential, and how is spatial profiling achieved? In this blog post, we will cover these questions with a focus on techniques that spatially resolve mRNA targets or the transcriptome.
Why spatially resolved transcriptomics matters
The adage "Location! Location! Location!" is just as valid for biology as it is for real estate.
Developmental biology is an excellent framework for understanding this concept. All the cells in the human body contain the same DNA. Yet, from this same genetic material, specialized cells emerge, forming very distinct tissues and organ structures as an organism develops (2). Nature, ever so clever, leverages carefully controlled gene expression gradients to activate the right development pathways at the right time (2–3).

Even after development, location influences cellular phenotype, state, and function because a cell's position relative to other surrounding cells dictates what signals it gets exposed to. These signals may come in the form of cell–cell interactions, soluble messengers, or even therapeutic molecules (2).
Devastating diseases emerge when the carefully coordinated interaction of cells within our bodies is disrupted. That is why gaining a deeper understanding of the biological mechanisms driving human development and disease requires careful assessment of how a cell's environment impacts its function or dysfunction.
Enter spatially resolved transcriptomics
The desire to assess cellular biology in a natural context is not new. In their review article titled “Museum of Spatial Transcriptomics,” Lambda Moses and Lior Pachter highlight radioactive in situ hybridization visualizing ribosomal RNA (1969) and globulin transcripts (1973) as prequel techniques to current-day spatial transcriptomics techniques (4). Although these early methods proved it was possible to spatially resolve biological molecules, they could only capture a limited number of targets at a low resolution.
Technological advancements in the 1990s and early 2000s lead to a boom in new transcriptomics methodologies and spatial analysis software (4). Much like the single cell sequencing field, only the academic institutions where early spatial profiling techniques were developed adopted the methods. However, with the commercialization of some of these techniques, spatially resolved transcriptomics has become more accessible to more researchers (2). From developmental biology to cancer, neuroscience to plant biology, and beyond, more publications come out everyday revealing a whole new world of biological insights.
Read more about some of our favorite spatial biology discoveries:
- Spatial transcriptomics publications you’ll want to read (Read here)
- Spatial transcriptomics publications you’ll want to read: 2022 edition (Read here)
How do you achieve spatial gene expression profiling?
Dozens of spatial transcriptomics techniques are currently available, which can make diving into the field daunting. A helpful way to orient yourself is to focus on the different ways spatial gene expression data is collected. Several recent reviews simplify the categorization of available techniques into two categories: sequencing-based spatial transcriptomics technologies and imaging-based spatial transcriptomics technologies (2, 5).
Sequencing-based spatial transcriptomics
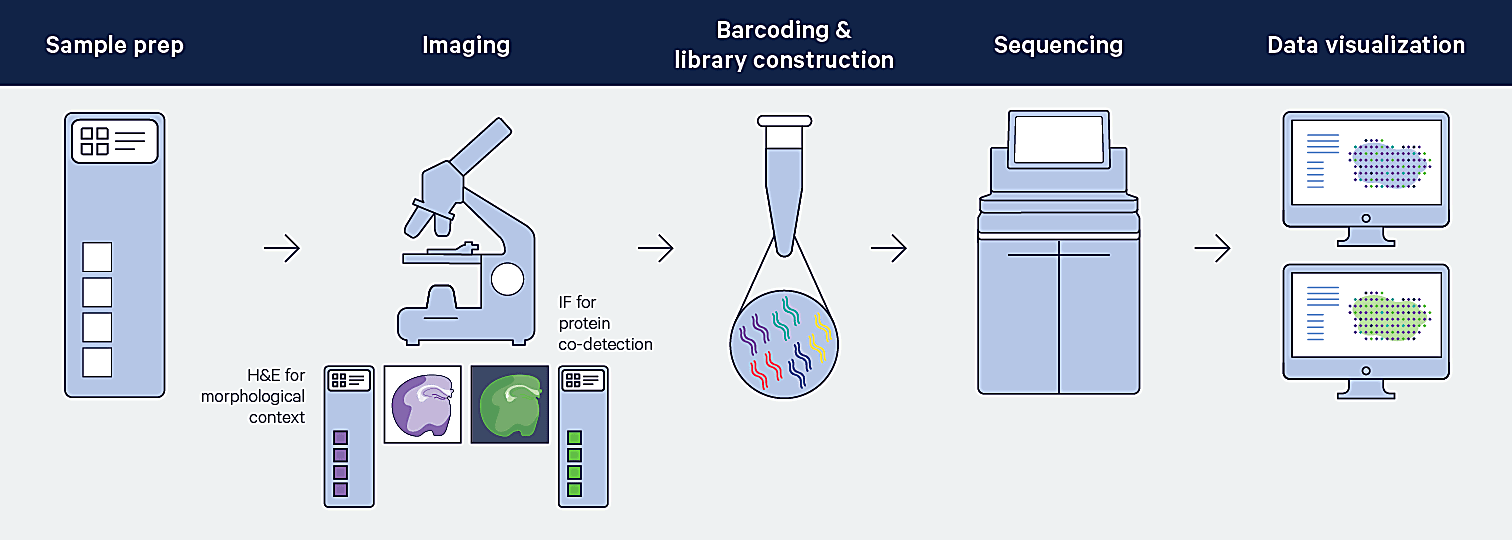
With sequencing-based spatial methods, mRNA is captured from tissues and then sequenced ex vivo (2, 5). You may be wondering: if sequencing occurs ex vivo, how is the spatial location of the mRNA target determined? Let’s take a look at how Visium Spatial captures the spatial context of profiled mRNAs.
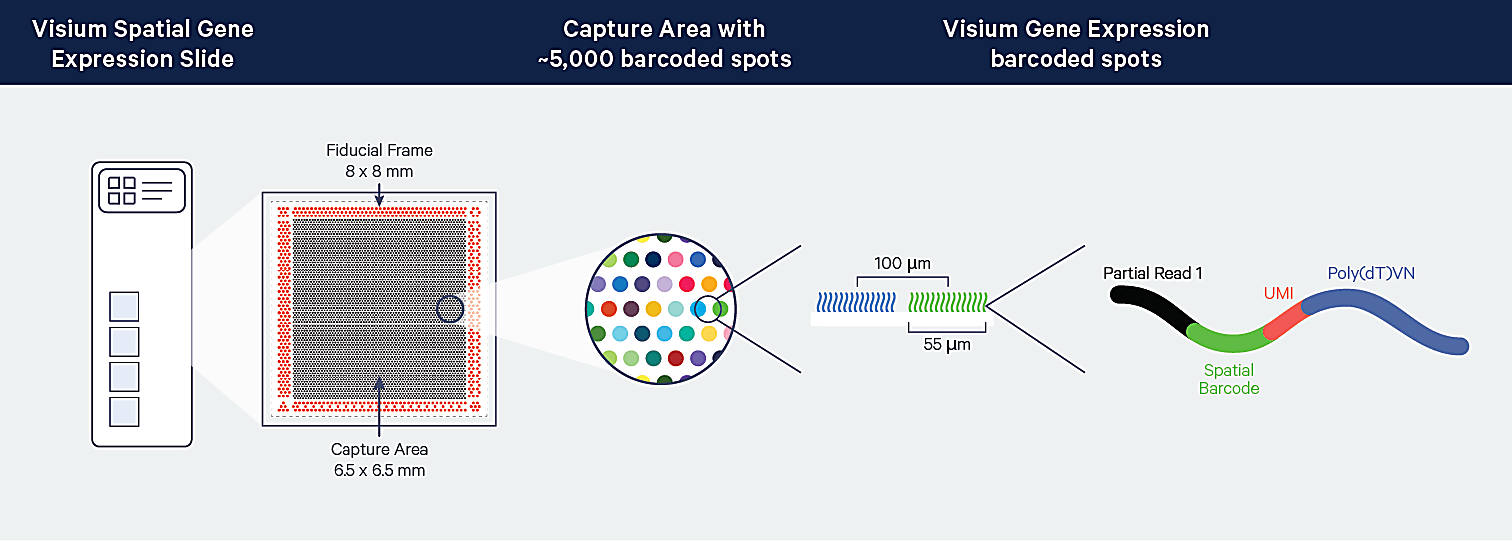
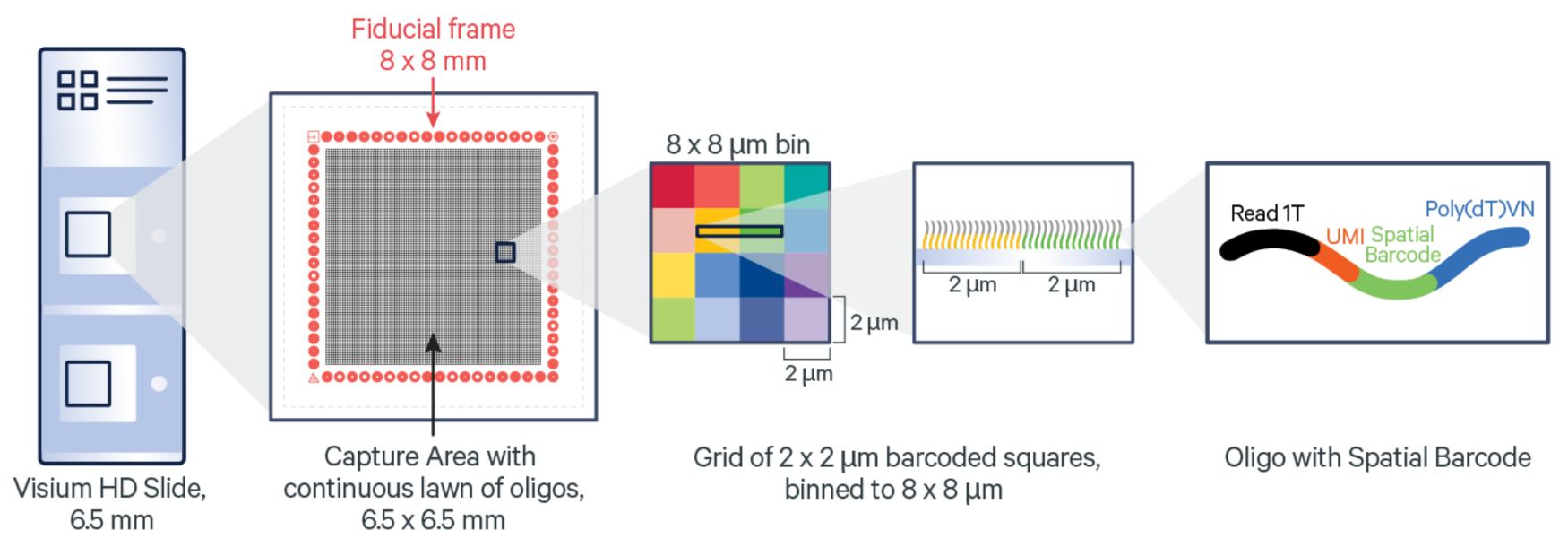
Visium Spatial leverages positionally barcoded, “array-based” capture of mRNA in which tissue sections are mounted on top of Capture Areas containing barcoded spots (6). For the fresh frozen (FF) tissue Visium assay, polyadenylated mRNA is captured. For the formalin-fixed paraffin-embedded (FFPE) tissue Visium assay, ligated probe pairs used for target identification are captured on the slide.
Complements of the spatial barcodes are incorporated during cDNA library generation (FF assay) or probe extension (FFPE assay), preserving spatial information. Barcoded libraries analyzed by next-generation sequencing are mapped back to a specific spot on the Capture Area.
Imaging-based spatial transcriptomics
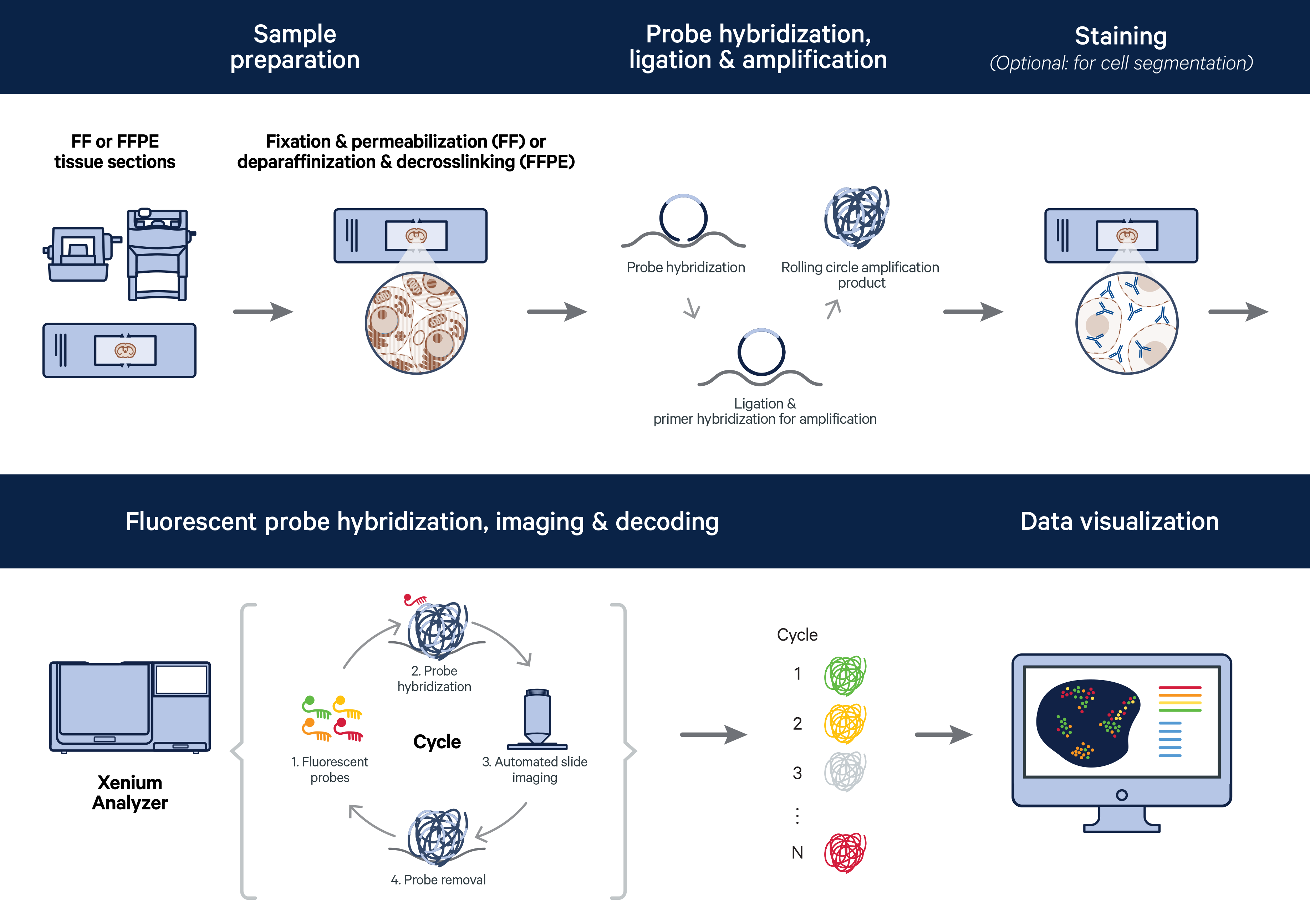
With imaging-based spatial methods, also referred to as microscopy-based spatial methods, mRNAs are imaged in situ (2, 5). This category is often further subdivided into in situ hybridization–based and in situ sequencing–based methods, which, despite the naming, does not rely on next generation sequencing (2, 7). Let’s focus on the imaging-based methodology leveraged in the Xenium In Situ platform to see how target identification and detection is achieved.
Circularizable padlock probes containing two regions that independently hybridize to the target and a gene-specific barcode sequence are used for mRNA labeling. Successful target recognition undergoes Xenium’s unique and highly specific four-step authentication chemistry. In the first two steps, both arms must stably hybridize to the target. In step three, a ligase joins together stably bound probe ends, generating a circularized probe. In the fourth step, successfully ligated probes are amplified using rolling circle amplification (RCA).
Detection is achieved with successive rounds of fluorescent probe hybridization to the RCA products, imaging, and probe removal. An optical signature specific to each gene is generated and a spatial map of the transcripts is built across the entire tissue section.
Which method is right for me?
There are many factors to consider when deciding between RNA sequencing–based and imaging-based spatial methods. A good starting point is to consider the experimental aim of your research with the recommendation of using sequencing-based methods for hypothesis generation and imaging-based methods for hypothesis testing (2). The basis of this recommendation is the level of information each method provides.
For example, sequencing-based Visium Spatial leverages unbiased whole transcriptome analysis, providing information on tens of thousands of genes, and therefore requires no a priori knowledge of the biology being studied. On the other hand, imaging-based Xenium In Situ is designed to profile hundreds to thousands of genes at high resolution, specificity, and sensitivity. This more targeted approach is ideal for taking a deep dive into targets of interest previously identified by other methods, such as single cell sequencing or unbiased spatial transcriptomics. It is important to note, however, that this does not mean that imaging-based spatial profiling is unable to discover new biology that generates hypotheses, as demonstrated in our blog post Behind the preprint: Combining single cell, spatial, and in situ for deeper biological understanding.
Taking your research spatial
The rise of spatial biology adoption is helping pave new avenues for basic and clinical research to expand our understanding of development and disease. Discover the possibilities that these technologies will open up to you by visiting our Visium Discovery Hub or interactively explore Xenium datasets from human breast or mouse brain tissue.
References:
- Marx V. Method of the Year: spatially resolved transcriptomics. Nat Methods. 18: 9–14 (2021). doi: 10.1038/s41592-020-01033-y
- Williams CG, et al. An introduction to spatial transcriptomics for biomedical research. Genome Med. 14: 68 (2022). doi: 10.1186/s13073-022-01075-1
- Asp M, Bergenstråhle J & Lundeberg J. Spatially resolved transcriptomes—next generation tools for tissue exploration. Bioessays. 42: e1900221 (2020). doi: 10.1002/bies.201900221
- Moses L & Pachter L. Museum of spatial transcriptomics. Nat Methods. 19: 534–546 (2022). doi: 10.1038/s41592-022-01409-2
- Method of the Year 2020: spatially resolved transcriptomics. Nat Methods. 18: 1 (2021). doi: 10.1038/s41592-020-01042-x
- Stahl PL, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 353: 78–82 (2016). doi: 10.1126/science.aaf2403
- Zhuang X. Spatially resolved single-cell genomics and transcriptomics by imaging. Nat Methods. 18: 18–22 (2021). doi: 10.1038/s41592-020-01037-8
