FFPE Human Ovarian Cancer with 5K Human Pan Tissue and Pathways Panel plus 100 Custom Genes
In Situ Gene Expression dataset analyzed using Xenium Onboard Analysis 3.0.0
Learn about Xenium analysis
Overview
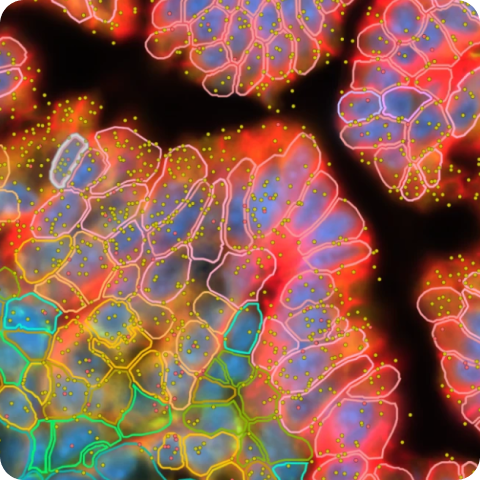
Xenium Prime 5K In Situ Gene Expression with Cell Segmentation data for human ovarian cancer (FFPE) using the Xenium Prime 5K Human Pan Tissue and Pathways Panel plus 100 Custom Genes.
relabel pipeline to update the output files to the XOA v3.2 formats (see release notes for details) for use with Xenium Explorer 3.2 features. The raw transcript, image, and cell segmentation data are unchanged.How to view data
Interactively explore data with Xenium Explorer by downloading the Xenium Output Bundle (or Xenium Explorer subset) file. The subset bundle contains the experiment.xenium, gene_panel.json, morphology_focus/ directory of multi-file OME-TIFF files, analysis_summary.html, cells.zarr.zip, cell_feature_matrix.zarr.zip, transcripts.zarr.zip, and analysis.zarr.zip files.
See the Getting Started with Xenium Explorer page for more details. Follow these instructions to view the post-Xenium H&E image or image alignment file in Xenium Explorer.
Biomaterials
FFPE-preserved tissue blocks were purchased from Discovery Life Sciences (Ovarian Papillary Serous Carcinoma: III-B (T3b N0 MX)).
Tissue preparation
Tissues were prepared following the Xenium In Situ for FFPE - Tissue Preparation Handbook (CG000578).
Probe hybridization, washing, ligation, amplification, and cell segmentation staining were performed following the Xenium Prime In Situ Gene Expression with optional Cell Segmentation Staining User Guide (CG000760).
Post-instrument processing followed the Demonstrated Protocol Xenium In Situ Gene Expression - Post-Xenium Analyzer H&E Staining (CG000613).
Gene panels
The Xenium Prime 5K Human Pan Tissue and Pathways Panel was designed to enable comprehensive cell type and cell state identification using publicly available single cell RNA sequencing data. The panel also covers canonical signaling pathways, as well as genes relevant to developmental biology, immuno-oncology, and genes that are well known in biomedical literature.
The 100 gene custom panel was selected based on differentially expressed genes from serial section matched Flex data from four tissue types: ovarian, cervical, breast, and uterine cancers. In addition, HPV16/18 probes were included for cervical cancer (based on information from the biobank where the FFPE tissue blocks were sourced).
Xenium Analyzer
The instrument run was performed following the Xenium Analyzer User Guide CG000584. The on-instrument analysis was run with Xenium Onboard Analysis v3.0.0.
| Metric | Ovarian cancer |
|---|---|
| Median transcripts per cell | 178 |
| Cells detected | 407,124 |
| Nuclear transcripts per 100 µm² | 385.3 |
| High quality decoded transcripts detected | 120,455,566 |
| Region area (µm²) | 86,812,984.2 |
Custom Cell Groups
Xenium data was imported into Seurat v5.0.1.9007, without transcript data, according to this vignette. Data was subsampled to 200k cells with Sketch. Using the FindClusters function (resolution = 0.6), 18 cell clusters were identified. One cluster was subclustered to separate fallopian tube epithelium and ovarian granulosa cells. Another cluster was subclustered to separate pericytes and tumor-associated endothelial cells. Annotation was performed using a variety of methods including gene set enrichment analysis (Enrichr), the Human Protein Atlas, literature searches, and comparisons to the matched Single Cell Gene Expression Flex data. Using the FetchData function, barcodes for each cluster were exported into a CSV and then imported into Xenium Explorer by creating custom cell groups. The annotation CSV export process is described in more detail in this Analysis Guide.
Custom Gene Groups
Using the Seurat object described above, we employed FindAllMarkers to identify top differentially expressed genes distinguishing each cell group. All genes are mutually exclusive to a single cell type with the exception of DSC3, which marks both granulosa cells and malignant cells lining cyst.
Pathology Annotations
Pathology annotations in the Annotated Post-Xenium H&E Image (OME-TIFF) file were created in QuPath v0.5.1 and converted to ome.tif format following this KB article. The annotation colors on the H&E image are: red = tumor, black = necrosis, blue = smooth muscle, dark green = fallopian tube, and light green = ovary.
Pathology annotations can be imported in GeoJSON format in Xenium Explorer v4.1 and later (see Annotation Layer documentation for details). The supplemental GeoJSON annotation file is aligned to the post-Xenium H&E image.
This dataset is licensed under the Creative Commons Attribution 4.0 International (CC BY 4.0) license. 10x citation guidelines available here.
