Visium HD Spatial Gene Expression Library, Human Colorectal Cancer (FFPE)
HD Spatial Gene Expression dataset analyzed using Space Ranger 3.0.0
Learn about Visium analysis
Overview
This dataset is associated with the tutorial Mapping the Tumor Microenvironment with Visium HD and Loupe Browser. Following this tutorial, you can learn how to:
- Review unsupervised clustering results
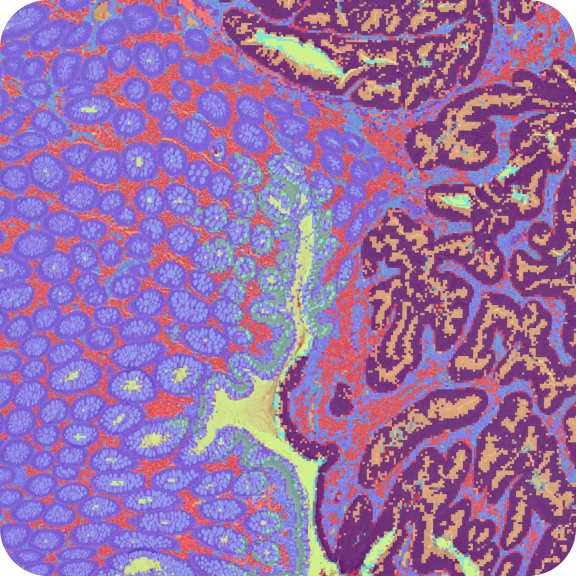
- Use goblet cell markers and the microscope image to visually assess transcript localization
- Map marker genes for tumor cells, cancer-associated fibroblasts, macrophages, and neutrophils to get a closer look at the tumor microenvironment
- Create a new group using known marker genes and perform differential gene expression to discover new markers
- Plot co-expression of tumor and fibroblast genes using Loupe’s new feature
This dataset was generated using late stage in-development protocols, reagents, consumables, and instrument software. However, limited changes were made between this version and the final product, and we expect final data quality to be equivalent.
Biomaterials
A human colon cancer sample (FFPE) was obtained from Discovery Life Sciences.
- Colon region: Sigmoid
- Stage: Unknown
- Sex: Male
- Age: 60
Sample preparation
A 5 µm section was taken from the FFPE tissue block with a microtome (Epredia HM355S). Sectioning, deparaffinization, H&E staining and imaging followed the Visium HD FFPE Tissue Preparation Handbook (CG000684).
Assay workflow
This dataset was generated using an in-development Visium HD Spatial Gene Expression protocol. Probe hybridization, probe ligation, slide preparation, probe release, extension, library construction, and sequencing followed the Visium HD Spatial Gene Expression Reagent Kits User Guide (CG000685).
- Slide serial number: H1-VM2JXXK
- Area: A1
- Instrument: Visium CytAssist
- Probe set: Visium Human Transcriptome Probe Set v2.0
Sequencing
- Sequencing instrument: Illumina NovaSeq 6000
- Sequencing configuration: 43 bp read 1, 50 bp read 2, 10 bp i7 sample index, and 10 bp i5 sample index
Analysis
Space Ranger v3.0 was used to map FASTQ files to the reference, detect tissue, align the data to the microscope and CytAssist images, and output feature-barcode matrices for further analysis.
Note that to process the FASTQ and image files using spaceranger count, the slide layout file must be downloaded and the --slidefile option must be used.
How to view data
To get started, download Loupe Browser v8.0 to explore the Loupe file, or read more about the other Space Ranger outputs.
This dataset is licensed under the Creative Commons Attribution 4.0 International (CC BY 4.0) license. 10x citation guidelines available here.
