When spatial gene expression meets FFPE tissue blocks: A modern-day love story
Though FFPE processing has revolutionized life science research, it damages RNA molecules, complicating transcriptomic-level investigations. Imagine bringing the comprehensive insights of spatially resolved transcriptomics to biobanked samples. Read this blog post to learn about the power of our newest offering, Visium Spatial Gene Expression for FFPE.
Much like Kodak film allows us to fix cherished moments of our lives in time, formalin fixing and paraffin embedding tissue lets us preserve precious biological samples. It is estimated that more than one billion tissue samples are stored in hospital and biobank archives, the majority formalin-fixed and paraffin-embedded (FFPE; 1), waiting for their biological secrets to be unlocked.
Since being first described over 100 years ago (1), FFPE tissues have become a vital resource for studying disease processes and aiding the diagnosis and treatment of many diseases. For example, in oncology, researchers and physicians rely on FFPE tissues to detect cancer genes as well as look for and track changes in tumor morphology over time (2). In pathology, FFPE tissue blocks are a gold standard for diagnostics.
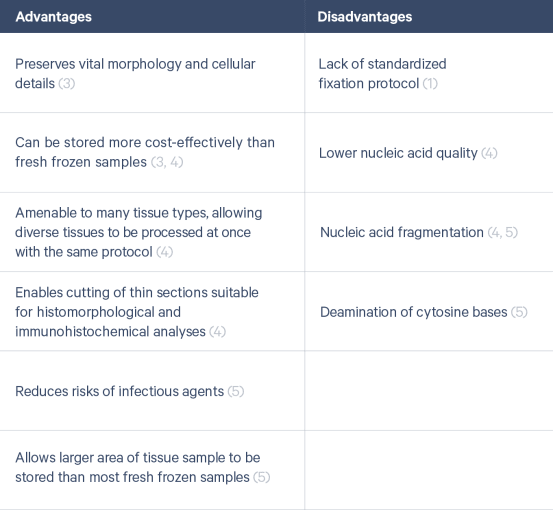
In recent years, spatially resolved transcriptomics has started to gain popularity for the spatial profiling of tissue samples allowing scientists to measure gene activity and map where it occurs. While FFPE processing is prized for its advantages, it is also associated with challenges that complicate sequencing-based approaches, causing some to favor fresh frozen tissues for these applications (Table 1). However, as technologies advance and are able to overcome these challenges, we can open the biobank vaults and bring transcriptomic-level analysis to archived samples.
Introducing Visium Spatial Gene Expression for FFPE
The new Visium Spatial Gene Expression for FFPE assay not only enables spatial gene expression analysis in FFPE tissues with the discovery power and throughput of RNA-sequencing but also pairs it with histological analyses already routinely done on these samples. Visium’s whole transcriptome analysis allows researchers to achieve true discovery in a morphological context. Furthermore, the platform removes analysis boundaries associated with predefined regions of interest by enabling the analysis of entire tissue sections so you don’t miss out on important biology. Altogether, this means you can determine which genes are expressed in your FFPE tissue, and where, as well as in what quantity.
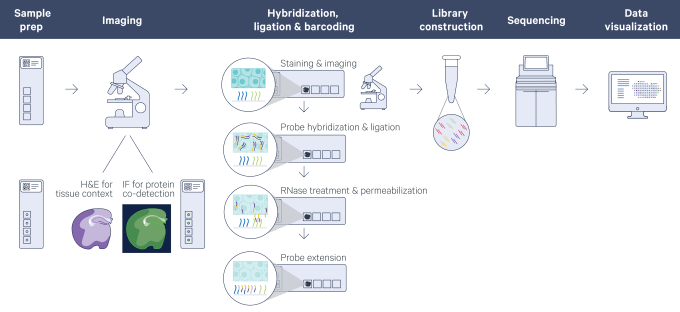
The spatially resolved gene expression data is subsequently layered over a high-resolution microscope image of the tissue section with the use of visualization software, making it possible to view the expression of any mRNA, or combination of mRNAs, within the morphology of the tissue.
FFPE research dreams are made of this
What challenges could we solve and what breakthroughs could we make with whole transcriptome spatial gene expression of FFPE tissue blocks? We asked a few leading translational researchers just that. Here’s what they had to say:
It is a genome wide technology. I can look at everything at the same time. And I think, that’s why it’s leveraging not only the cancer field but the study of other complex diseases as well.
- Luciane Kagohara, Instructor of Oncology, Johns Hopkins University
It's the leading technology, in my mind. To really help us to further improve on our therapies, to identify new therapies to test in patients, and to really move precision oncology so that it's truly patient-specific.
- Elizabeth Jaffee, Deputy Director, Sidney Kimmel Cancer Center, Johns Hopkins University
By having Visium accessible in FFPE samples, we're now really able to unlock just tons of legacy data. I can go back to every failed trial that ever was and try to understand now why it didn't work to see if we can uncover new mechanisms for treatment. And that's just thrilling.
- Elana Fertig, Assistant Director, Research Program in Quantitative Sciences, Johns Hopkins University
These are just a few of the exciting possibilities, and we can’t wait to see what you will discover!
Interested in learning more about Visium Spatial Gene Expression for FFPE? Join us for a series of webinars that will introduce the technology and cover getting started with the workflow, sample preparation, data analysis and visualization, and more.
Register now | Americas and Europe →
References
- Blow N. Tissue issues. Nature 448, 959–960 (2007). doi: 10.1038/448959a
- Sim J. (2018, June 20). What are FFPE samples. Retrieved April 20, 2021, from https://www.geneticistinc.com/blog/ffpe-samples (no longer available)
- Kokkat TJ, et al. Archived formalin-fixed paraffin-embedded (FFPE) blocks: A valuable underexploited resource for extraction of DNA, RNA, and protein. Biopreserv Biobank 11(2): 101–106, 2013. doi: 10.1089/bio.2012.0052
- Mathieson W and Thomas GA. Why formalin-fixed, paraffin-embedded biospecimens must be used in genomic medicine: An evidence-based review and conclusion. J Histochem Cytochem 68(8): 543–552, 2020. doi: 10.1369/0022155420945050
- Mathieson W and Thomas G. Using FFPE tissue in genomic analyses: Advantages, disadvantages and the role of biospecimen science. Curr Pathobiol Rep 7: 35–40, 2019. doi: 10.1007/s40139-019-00194-6
About the author:

