Improved de novo assembly and variant calling with new Chromium Software
We are excited to introduce the latest evolution in 10x Chromium Software, Supernova 1.1, Long Ranger 2.1 and Loupe 2.1. Packed with new features and enhancements, the latest software versions offer significant improvements for de novo assembly, variant calling, linked read data visualization and usability. Check out the highlights below.
Improved Assembly Quality and Reduced Resource Requirements
Supernova 1.1 - Download and Release Notes
- Assemble 10-20% longer contigs with new algorithm enhancements
- Optimize resource utilization with new resource management controls and a 25% reduction in memory requirements for human assembly
Dramatically Increased Sensitivity for SNPs, indels and SVs
Long Ranger 2.1 - Download and Release Notes
- Confidently call variants with a substantial increase in SNP and indel sensitivity using a new haploid calling approach
- Discover up to 30% more novel SNP calls for WGS and up to 85% more for WES with Lariat aligner enhancements
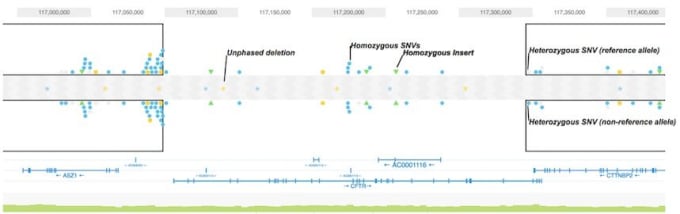
- Identify 1000’s of 50bp-30kb heterozygous and homozygous deletions utilizing the new haplotype-aware mid-scale SV caller
- Confidently identify large deletions with an increase in >30kb deletion sensitivity using a revamped, unified large-scale SV caller
- Greater reference flexibility with support for GRCh38 and expanded support for non-human references
A Clearer View of Variants and Haplotypes
Loupe 2.1 - Download and Release Notes
- Easily see the big picture with new visualization of global phase block coverage for the entire genome
- Quickly focus in on phased variant types using the redesigned haplotype view
Additional Improvements
- Streamlined data analysis with simpler integration into existing workflows and better QC support using a new demultiplexing pipeline
- New and updated datasets for NA12878 and HCC1954 tumor-normal pair for both WGS and WES updated for Long Ranger/Loupe 2.1
- Seven new Supernova datasets from upcoming publication
- Updated GIAB and 1K Genomes datasets
Detailed information, including documentation and system requirements, may be found on the 10x Support Site. We strive to make our software as valuable and easy-to-use as possible, so please do not hesitate to contact us with questions or feedback at support@10xgenomics.com.
