Immune repertoire profiling at single cell resolution
We are excited to announce the launch of our new Chromium™ Single Cell V(D)J Solution. It is the first high-throughput solution for profiling paired V(D)J transcripts from hundreds to millions of lymphocytes and expands our industry-leading single-cell product portfolio. We think the ability to more completely identify and characterize the body’s immune agents (T and B cells) at the single-cell level will impact our understanding and development of more effective and durable immunotherapies, including immuno-oncology (I/O) drugs for targeted cancer therapy. We're going to be introducing this new application at the annual meeting of the American Association for Cancer Research (AACR) in Washington, DC, April 1-5 at Booth 1644 and in our workshop on Sunday, April 2.
The Chromium Single Cell V(D)J Solution enables assembly of full-length V(D)J sequences —5’ untranslated region (UTR) to constant regions— on a cell-by-cell basis, providing high resolution insights into the adaptive immune system. Five-prime barcoding limits bias caused by complex multiplex PCR, enables the detection of germline and somatic variants across the entire V(D)J segment and supports cell type classification or phenotyping.
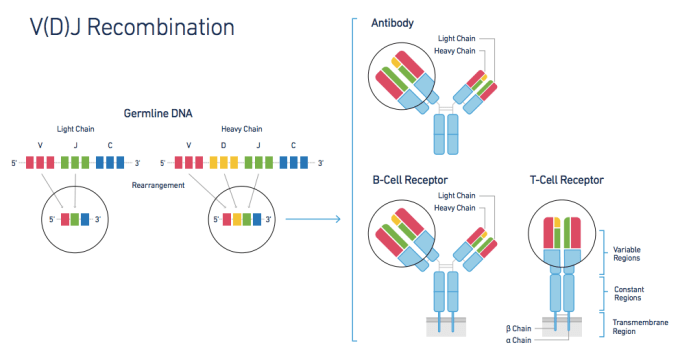
Antigen specificity and immune system diversity are generated by V(D)J recombination and somatic hypermutation in a clonal manner. Antigen specificity is determined by two co-expressed genes, the heavy and light chains of the B-cell receptor and the alpha and beta chains of the T-cell receptor. Single-cell sequencing reveals the true clonality and diversity of the immune repertoire. This immune repertoire sequencing allows for a detailed analysis of the diverse range of antigen receptors, enhancing our understanding of immune system complexity and functionality.
T and B cells generate the incredible diversity of heterodimeric cell surface and secreted antigen receptor genes through the somatic recombination of Variable (V), Diversity (D) and Joining (J) gene sequences. Accurate characterization of this repertoire of paired T-cell receptor alpha and beta chain genes is critical to the characterization of the interaction between antigens and T cells. This understanding will enable researchers to pursue next-generation studies into the true diversity of the paired T-cell and B-cell receptor repertoire. Further, it will allow researchers to apply this understanding to areas such as vaccine development, clonal immune cell dynamics, immune responses to checkpoint blockades, and the development of recombinant antibodies and engineered T cells used in immunotherapies for cancer and other diseases.
The Chromium Single Cell V(D)J Solution Features:
- High cell capture efficiency up to 65%
- Scalable throughput from 100 cells to millions of cells
- Supports lymphocytes, PBMCs, cell lines, FACs-isolated cells and MACS® MicroBead-enriched cells
- Low doublet rate: 0.9% per 1,000 cells
- Compatible with Illumina® HiSeq® 4000/2500/NextSeq®/ MiSeq® sequencers
- Partition 100-10,000+ cells per channel in < 7 minutes
- Run 1 to 8 channels in parallel
The new, comprehensive solution, designed, developed and manufactured solely by 10x Genomics, can be run on the Chromium™ Controller, as well as the dedicated Chromium™ Single Cell Controller.
Find out more about the new Chromium Single Cell V(D)J Solution here. And, for even more information, check out the full press release.
