How TGen drastically reduced Xenium per-sample costs for large cohorts with tumor microarrays
“The combination of the FFPE compatibility, as well as the high throughput of the Xenium system, helps us start to envision how we might really do these projects at scale. What I mean is, instead of just ten or fifteen samples, we could start thinking about hundreds of samples that we could process.” – Nick Banovich, PhD
It’s a familiar story in science: a new, cutting-edge technology is released. It promises deeper insights and more information, but with the catch that it’s expensive and/or low-throughput. So a technology that could help quickly revolutionize a field is stuck behind a paywall until economies of scale make it more affordable for larger studies and more users.
However, Nick Banovich, PhD (The Translational Genomics Research Institute (TGen), part of City of Hope), an early adopter of Xenium, had a question: what if you could optimize not only per-sample cost, but do so in a way that made multi-sample studies not just feasible, but logical?
We recently had the opportunity to sit down with him and discuss how he answered that question. Read on to learn how he developed a streamlined tissue microarray (TMA) protocol that enabled an up-to-eight-fold reduction in the per-sample cost of Xenium.
Starting a little bit more broadly—would you mind describing both your research and your lab at TGen?
One of my roles at TGen is to operate our spatial transcriptomics technology core. The core offers Xenium and Visium HD as services to both internal investigators at TGen as well as external investigators outside of our institute.
My primary function is my research lab, which is focused on understanding how gene regulatory changes impact disease outcomes. Over the past several years, we've built our foundation on single cell transcriptomics and using this to understand the molecular dysregulation of disease with cell-type resolution.

One of our major projects is focused on pulmonary fibrosis. We also focus on correlative analyses from patients undergoing CAR T-cell therapies for brain tumors. So we use very different biological systems, but they’re unified by the set of tools and approaches, i.e. looking at molecular dysregulation in disease at cell-type resolution.
Over the past couple of years we've started to put more of a focus on spatial technologies, and we've been using Xenium since February of 2023. We think about the same types of concepts that we thought about in single cell, but obviously spatial data has a lot of benefits: understanding not just the cell-type-level dysregulation, but the organization of cells into architectural niches.
Can you tell us a bit more about your TMA approach and why it’s a big deal for cost reductions?
One of the things that's unique about Xenium, and unique about imaging-based spatial transcriptomics, is that it's the first time in genomics, since the advent of next generation sequencing, where we're not doing an assay and then putting that on a sequencer. It's really an all-in-one platform.
Because of that, our cost drops linearly as we add more samples to a single run. We were early adopters of single cell multiplexing approaches. Those drop the front-end cost, but the sequencing costs increase with greater numbers of cells. With Xenium, it costs the same to analyze 10% of the available sample area as it does to analyze 100% of the available tissue area, so there’s a true linear cost drop.
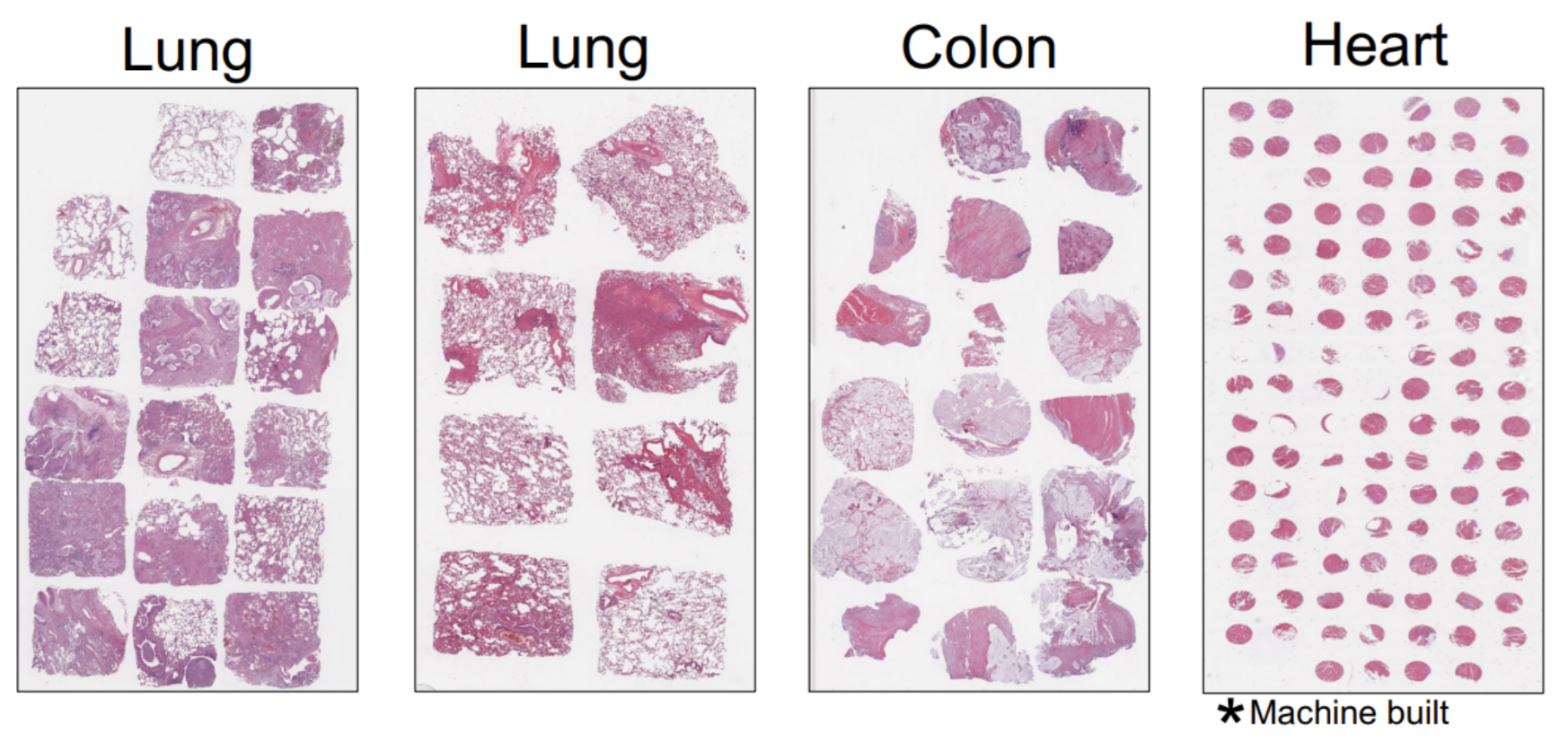
So we really thought about how we could maximize this area, and we realized that you could build a TMA and place multiple samples on a single Xenium slide. After setting up these TMAs, we realized you’d get about 20% more area per sample if you used a square biopsy punch versus a circle biopsy punch, so we had a square biopsy punch fabricated (Figure 2).
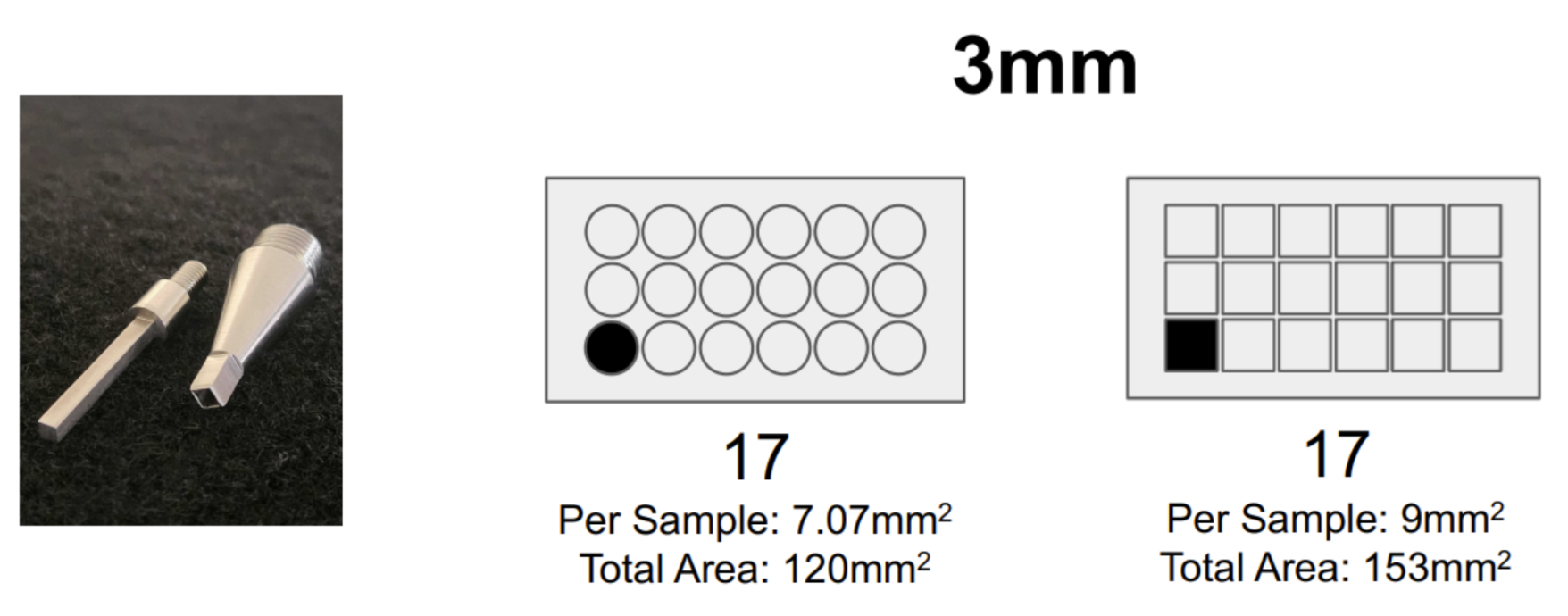
That's fascinating. And it's not often that biologists get to spend time in machine shops!
Luckily, Arizona State University actually has a die-cast program, so we were able to work together with them. We brought them a commercial hand punch that had a variety of different circular biopsy punches, so we were able to give them the handle of that unit and they fabricated the square punch to fit into it.
We've opted for the square punch because we're maximizing area. But we've also had some collaborators who use one of the older machine punches since they’re focused on getting much smaller punches. We had a collaborator who built a tissue array using 0.6mm biopsy punches that they created. So it's extremely flexible.
Considering the possibilities these tissue microarrays hold. How has this approach helped your own work? And what do the cost savings look like?
We’re definitely doing some large studies. Right now, we’re profiling samples from patients with various types of lung disease, mostly pulmonary fibrosis.
As far as cost savings go: let’s say you’re running an experiment with 30 samples (Figure 4). Let’s estimate $9,000 for a run on Xenium, which is two slides of up to 237 mm2 of tissue each, and we’ll assume two sections on a slide. The cost also includes a Xenium panel, as well as reagents and consumables. That’s $68,000 for those samples, or a per-sample cost of ~$2,300, which might be somebody’s entire supply budget on a grant for the year.
Now switch to the tissue microarray format and you can fit all those samples on two slides (Figure 3), which is one run on Xenium. You go from $68,000 and multiple runs to analyze those samples, to $9,000 and one run. You go from $2,300 per sample, to $300 per sample.
We really think of this as enabling researchers to do projects at this scale that are feasible to propose to the NIH, rather than pilot studies with two or three samples.

The great thing is, this impacts both ends of the spectrum. It impacts people like me, who have already bought and invested into this technology and are doing huge projects. It also really impacts people who are at the early stage of their spatial journey by letting them get some good preliminary data for a grant, or even write a short paper, and leverage that to start going bigger.
So what are the trade-offs?
This small punch is a good representation of all of the architectural features I’d expect to see in a normal, healthy lung sample. If we look at a broader sample, you're kind of just resampling the same types of structures over and over again, or you can choose to sample a representative region.
There's definitely a trade-off on the area you want to image versus the number of samples you want to image. We found that, for us, a punch actually gives us quite a bit of tissue heterogeneity. Here's an example of a punch (Figure 4):
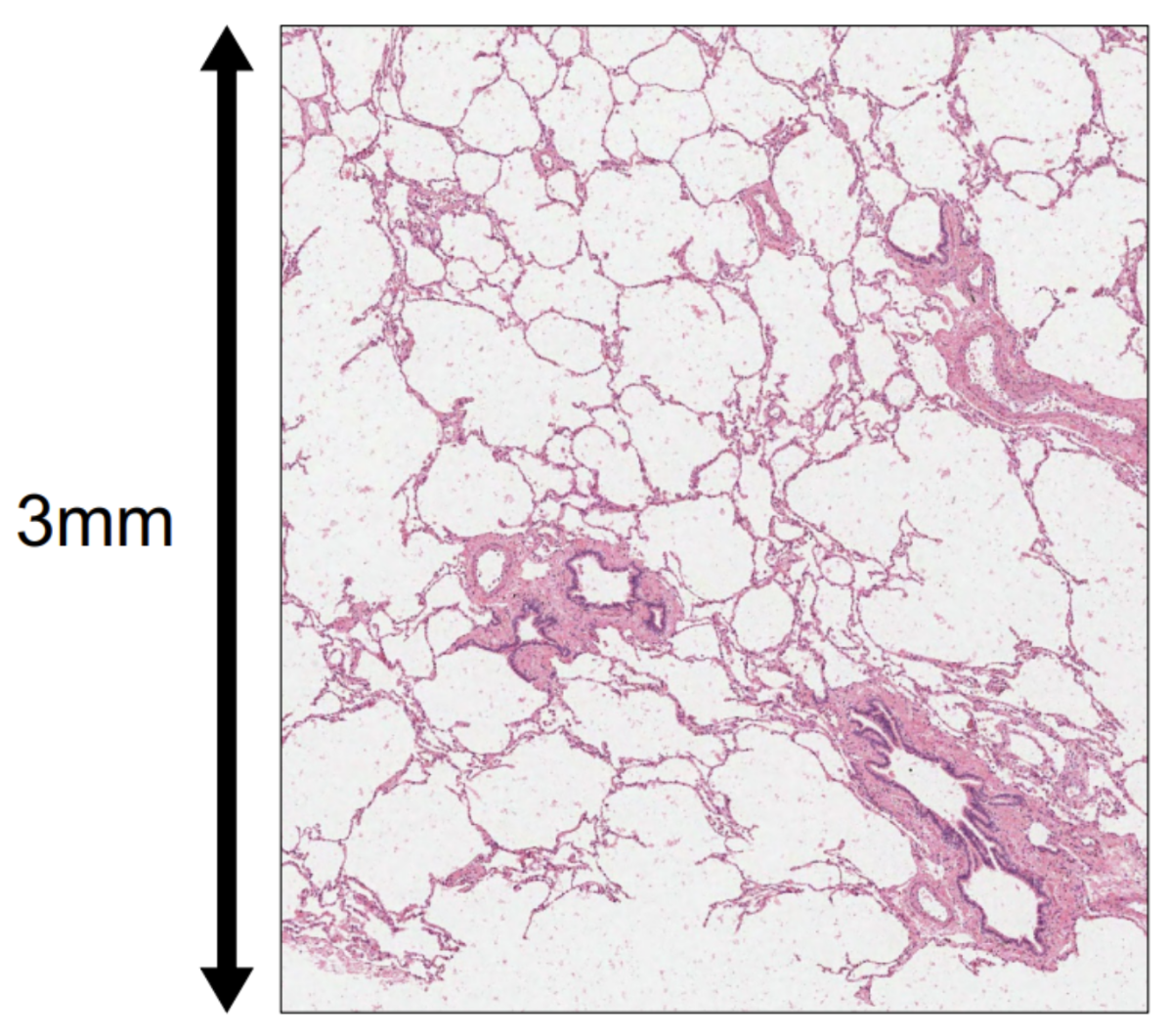
You can see it captures alveolar structures, a couple of small airways, and vessels. This small punch is a good representation of all of the architectural features I’d expect to see in a normal, healthy lung sample. If we look at a broader sample, you're kind of just resampling the same types of structures over and over again, or you can choose to sample a representative region.
Yes, this reduces the representation, but—barring model organism organs—you’re never going to be able to fit the entire tissue section on the slide. If you're dealing with human tissue, whether that's tumors, lungs, hearts, or livers, you're never looking at the whole tissue anyways. You're always picking something that's representative.
So I think there are two questions people need to ask themselves. One, how much area do they need to see the structures they care about? And two, do they care more about sampling large regions because they have a lot of rare, interesting biology, or do they want to look at lots of samples to learn principles of this tissue type or disease type?
Our lab has taken the second approach. We're going to get a good representation of all of the features of disease across all of those individuals. Even if we have less power to say something about an individual, it will give us a lot of power to say something about the biology of lung disease at large.
So it’s analogous to the early days of sequencing where researchers had to balance breadth versus depth.
Exactly. There's certainly times where, for example, we've got samples that are from rare diseases or samples from really unique patients that it makes sense to spend more “real estate” on. The beauty is, this system is flexible: you can do most of your study using tissue microarrays, use that to inform which patients may be interesting to focus on, then go back and deeply profile that subset.
Can we get some insight into the complexity and how difficult it is to set up this TMA process?
Complexity is in the eye of the beholder. I'd say this process is pretty easy: the off-the-shelf tissue punch kit, which is a circular punch, comes with molds to make the receiving blocks as well as different punch sizes.You need a microtome for sectioning. If you're not starting with tissue samples from pathology, you need the equipment to embed your tissue.
But the tissue microarray process is actually quite straightforward. If you use this kind of configuration, you can start to get into square punches, and we designed our tissue microarray configuration to be maximized for the Xenium imageable area.
All this takes a bit of setup to get going. If you just want to take advantage of the tissue microarrays and don't need to fully optimize every single piece of the process, it’s very easy to get started.
If I were an investigator who was interested in this, I could just go to TGen, right? I could just send you my tissue blocks or whatever you need, since you’re doing the tissue punches and turning these TMAs around routinely. Is that fair to say?
Correct. I think a lot of our external users are in a situation where the core services that could help them accomplish these studies are kind of disjointed. Maybe there's a pathology core that embeds the samples, then a histology core that does sectioning, then a genomics core that does Xenium.
It’s difficult to coordinate these things, so I’d say a lot of our users right now are coming to us because we have the ability to do any (or all) of this routinely, and they just want an all-in-one service.
So, speaking of your all-in-one service: if someone wants to collaborate with you and your lab, what’s the best way to reach out or get into contact?
One way is our website, which is TGen’s Center for Spatial MultiOmics (COSMO). We also have an e-mail address, which is cosmo@tgen.org, that will go straight to me.
You said in your earlier webinar with us that you’re an early adopter of these technologies. What made you opt for single cell spatial for your research, and what made you choose Xenium specifically?
The oldest sample we've profiled was about 15 years old. It had just sat on my collaborator’s desk in a shoebox that he touched and dug through with bare hands over the course of those years—everything that just makes us, as RNA people, die on the inside, you know? Except we were still able to get good data from it [with Xenium].
As we were looking at the competitive landscape, one of the things we really liked about Xenium was the ability to use FFPE tissues. A lot of our tissues are FFPE, and we liked that Xenium was able to still capture transcripts even from old, degraded FFPE samples that were collected without the intent of ever using them in a molecular assay.
The anecdote I like to use is that the oldest sample we've profiled was about 15 years old. It had just sat on my collaborator’s desk in a shoebox that he touched and dug through with bare hands over the course of those years—everything that just makes us, as RNA people, die on the inside, you know? Except we were still able to get good data from it. So I think Xenium has the ability to perform on those kinds of archival tissue samples.
This catapults us into the next part of our conversation—the ability to use FFPE samples in these tissue microarray approaches (Figure 1) is part of what really made us excited about the Xenium platform. The combination of the FFPE compatibility, as well as the high throughput of the Xenium system, helps us start to envision how we might really do these projects at scale. What I mean is, instead of just ten or fifteen samples, we could start thinking about hundreds of samples that we could process.
So, wrapping up: is there anything that's really cool or interesting that you want to tell us about that you haven’t gotten a chance to?
I think we've covered it! There's all sorts of cool biology we’re looking at, but every person's cool biology is going to be different, right? So the most exciting thing to me is to have more people thinking about these studies with bigger populations and really digging into their own biological questions with this kind of new dimension of information.
Fantastic; thank you for your time!
This was pretty fun! Thank you, too.
Interested in incorporating tumor microarrays into your single cell spatial imaging work? Check out TGen’s Center for Spatial MultiOmics (COSMO), or learn more about what Xenium can do for you here.
And learn more about costs savings on Xenium! Speak to your local 10x Genomics representative.
