Appreciating antibody diversity: unexpected isotypes found in response to primary infection
Scientists from the Walter Reed Army Institute of Research used single cell immune profiling to uncover a surprising role for plasmablasts secreting IgA, an antibody isotype typically found in mucosal membranes, in the immune response to primary infection by dengue virus (DENV), a mosquito-borne pathogen known to cause dengue fever. These results are surprising for more than one reason: IgA isn’t typically associated with infection recovery, and DENV has no known mucosal involvement, either during transmission or replication. Read more to learn how antibodies can protect us from viruses—but how researchers are still exploring which isotypes bear that responsibility, how they interact with virions to provide protection, and at what time points during infection.
When antibodies help viruses instead of us
Antibodies are fascinating, dynamic molecules, and crucial weapons in the adaptive immune system’s arsenal to target and clear pathogens. For example, when someone is infected with a virus, antibodies can target and coat the surface of the virus, essentially neutralizing it by preventing the virus from interacting with host-cell surface receptors that enable viral entry.
However, when it comes to viruses, antibodies’ protective functions can also serve as a double-edged sword. Depending on how they bind to the virus—including where they bind and how tightly—and how concentrated they are, certain antibodies target virions to host leukocytes with surface receptors that, in turn, bind to the antibodies. In most circumstances, these immune cells would destroy pathogenic particles via phagocytosis, but in this case, they could be infected with the virus. This phenomenon is called antibody-dependent enhancement (ADE) of infection. Among other infectious diseases, ADE has been widely observed in dengue fever. But researchers are still learning how the adaptive immune response to infection may work to drive dengue severity, particularly in secondary infection, which notoriously manifests in worse symptoms than an individual’s first encounter with the virus (1).
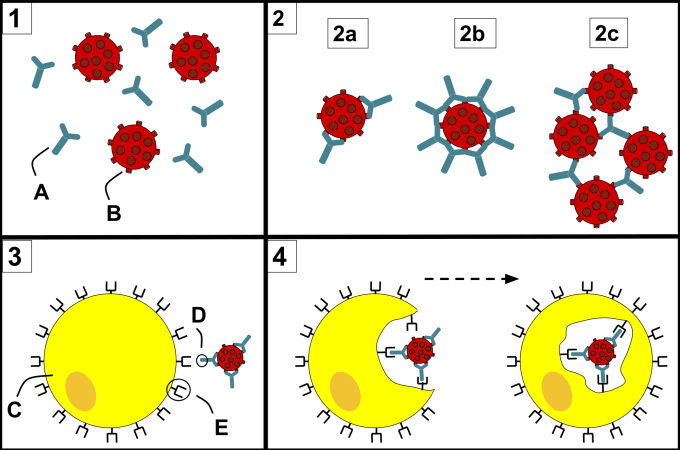
This duality in antibody function stems from antibody diversity. There are millions of B cells circulating in our bodies, each presenting immunoglobulin receptors—antibodies, when secreted—with specificity for a unique epitope, a small molecular surface on a pathogen or virus. However, because invading pathogens come in all shapes and sizes, some of these antibodies will be cross-reactive. They can bind non-specifically to more than one pathogen, and will be more or less effective than other antibodies at facilitating the neutralization or destruction of those pathogens. This risk makes understanding antibody diversity at detailed, molecular resolution all the more important, especially for efforts to discover novel neutralizing antibodies and develop effective therapies.
Antibody isotype diversity: how it plays out in viral protection
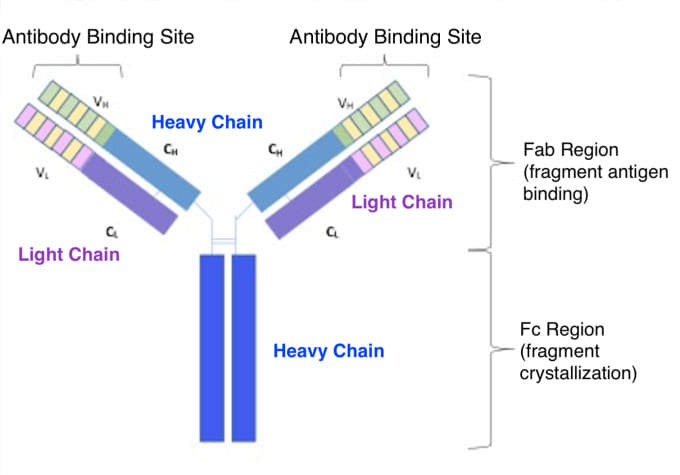
Beyond their innate variability in binding specificity and neutralization potency, antibodies have structural diversity that influences their function, prevalence, and location in the body. Antibodies are composed of amino acid molecules organized into two light chains and two heavy chains. At the tips of the antibody’s Y-shaped arms is a variable region that gives each antibody its unique specificity for a single antigen. The remaining sequences of the antibody are constant, and it’s the heavy chain components of this constant region that determine an antibody’s isotype. There are five isotypes, or immunoglobulin (Ig) classes, of human antibodies, including IgM, IgD, IgG, IgA, and IgE.
The composition of an antibody’s constant region, and therefore its isotype, is determined as the Ig gene undergoes recombination during B-cell development and maturation. Initially, immature B cells express the IgM antibody isotype, which is bound to the surface of the cell. As B cells mature, they express both IgM and IgD on the cell surface and are ready to respond to antigens. Once exposed to the right antigen, B cells are immunologically activated and begin to divide and differentiate into IgM- and IgD-secreting plasma cells. After this point of activation, and in response to further external activation by cytokines, the Ig gene encoding instructions for the constant heavy chains of the antibody will undergo rearrangement in select replicating daughter cells. This process is called immunoglobulin class switching and can lead to the secretion of other antibody isotypes, including IgE, IgA, or IgG.
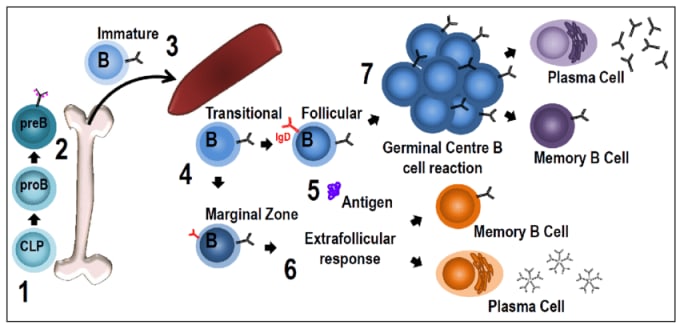
Though antibody isotypes derived from a single activated B cell have the same antigen specificity, they have distinct qualities and roles in the immune system. Some are more prevalent than others in serum; some can form complex multimer structures with greater avidity for pathogens; and some localize to the bloodstream or mucosal membranes. While immunologists continue to expand our knowledge of how the immune system works, antibody isotypes show us that there is a world of complexity still to be explored.
This complexity has more than theoretical implications. Researchers will need to characterize antibody diversity at molecular resolution to understand the immune response to infection and develop better treatments for global infectious diseases. What isotypes are prevalent in the immune response and during what time points in the process of infection recovery? What roles do different isotypes adopt in infection recovery, or, conversely, in adverse events like antibody-dependent enhancement of infection?
Answers are within reach, as outside-the-box research leverages innovative tools to gain deep insights into immune complexity. In the following study, learn how one group of researchers sought to uncover the molecular diversity of antibody-mediated humoral immunity resulting from infection by dengue virus (DENV), and how they used single cell immune profiling to explore the evolution of the immune repertoire upon primary and secondary infection.
Antibody diversity in primary and secondary DENV infection
Secondary infection by DENV is worse than primary infection, but researchers aren’t sure why. In their recent paper out in EBioMedicine, investigators from the Walter Reed Army Institute of Research speculated that this severe secondary infection may result from waning antibody-mediated cross-recognition of DENV particles. Or perhaps because IgG1 antibodies, known to be prevalent in secondary infection, are too good at opsonizing DENV—that is, guiding them to innate immune cells to be phagocytosed, but, in the process, errantly enhancing viral infection. Alternatively, the team speculated that something may have been overlooked in past studies of the immune response to DENV infection, since most studies focused entirely on secondary infection and restricted Ig receptor analysis to cells that expressed IgG or IgM only (2).
The team sought to fill this gap in knowledge of DENV by taking an unbiased approach to study the full diversity of plasmablast and antibody-mediated humoral immunity during primary and secondary DENV infection. Leveraging single cell immune profiling, they analyzed FACs-sorted CD19+ B cells from PBMCs of 6 pediatric patients with confirmed primary or secondary DENV infection. Out of the 9,027 B cells recovered, transcriptomic data revealed four distinct B-cell populations corresponding closely with the transcriptional profiles of naïve and memory B cells, plasmablasts, and pre-plasmablasts.
Subsequent analysis of antibody isotype diversity within these four cellular populations and between samples from patients with primary and secondary infection revealed that plasmablasts from individuals with primary infection had equal expression of IgA, IgG, and IgM. However, the team observed significant IgG1 bias within the plasmablast compartment of individuals with secondary DENV infection, and only modest representation of IgA or IgM (2). Further evidence of clonal expansion and somatic hypermutation in IgA class-switched plasmablasts in both primary and secondary infection suggested that these cells, and IgA, play an unexpected functional role in mediating the immune response to DENV.
With full-length, paired B-cell receptor sequence information from their single cell assay in hand, the team sought to determine the extent of that role. First, they measured how effectively IgA can bind with and neutralize DENV virions in comparison to other represented isotypes. They selected and synthesized 56 monoclonal antibodies (mAbs), a mix of candidates from both their primary and secondary infection samples that included IgM, IgG1, IgG3, and IgA1 isotypes. Leveraging a virus-capture ELISA method, they screened the antibodies for their ability to bind to DENV. As expected, they found a higher proportion of antibodies from secondary infection exhibiting DENV reactivity, including 76.6% of represented IgG1 mAbs. Unexpectedly, however, they noted that 47% of the mAbs synthesized from antibodies with an IgA isotype were DENV reactive (2).
Subsequent testing of antibody neutralization potency revealed more differences between the mAbs from primary and secondary infection samples. While only 37.5% of DENV-reactive mAbs from primary infection samples showed neutralization potential, 92% of reactive antibodies from secondary infection showed viral neutralization (2). These findings ultimately suggest that while mAbs from primary infection samples, including IgA, can react with DENV, this reactivity is general. In fact, the prominent IgA isotype in primary infection may result from cross-reactive, pre-existing memory B cells reawoken in response to primary DENV infection. The research team speculated further that the neutralization power of IgA might be more indirect. IgA antibodies may compete with IgG1 antibodies for binding to the virion. Though IgA doesn’t typically have the right heavy-chain structure to opsonize virions, binding competition could prevent antibody-dependent immune enhancement of DENV infection mediated by IgG antibodies.
While there’s more work to be done to fully understand the role of IgA in DENV infection recovery, this study shows the power of molecular-level resolution in uncovering the complex features of DENV-elicited humoral immunity. With the insights provided by single cell immune transcriptional profiling and receptor sequencing, researchers can see the immune response to infection at a whole new level of detail and take concrete action to develop effective treatments for infectious diseases.
Want to learn more about this study, conducted by scientists at the Walter Reed Army Institute of Research? Read their publication →
Find out how single cell immune profiling technology from 10x Genomics is advancing infectious disease research with these additional resources →
References:
- AL Rothman. Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nat. Rev. Immunol. 11, 532–543 (2011).
- AT Waickman et al., Transcriptional and clonal characterization of B cell plasmablast diversity following primary and secondary natural DENV infection. EBioMed. 54,102733 (2020).
