Answering your questions about Xenium In Situ: Spotlight on getting started and panel design
Diving into a new technology is exciting. However, there’s also a learning curve associated with getting familiar with new workflows and experimental design. That’s why we recently hosted a “Getting started with Xenium In Situ” webinar series, to make sure you have the information to help you be as successful as possible with your experiments.
In our first session, Ian Fiddes, PhD (Senior Manager, Computational Biology), and Albert Kim, PhD (Senior Scientist), walked through the Xenium workflow, Xenium panel offerings, and important considerations when designing custom panels. Keep reading to preview some highlights from the webinar’s Q&A section, and then watch the full webinar on-demand.
General assay and panel capabilities
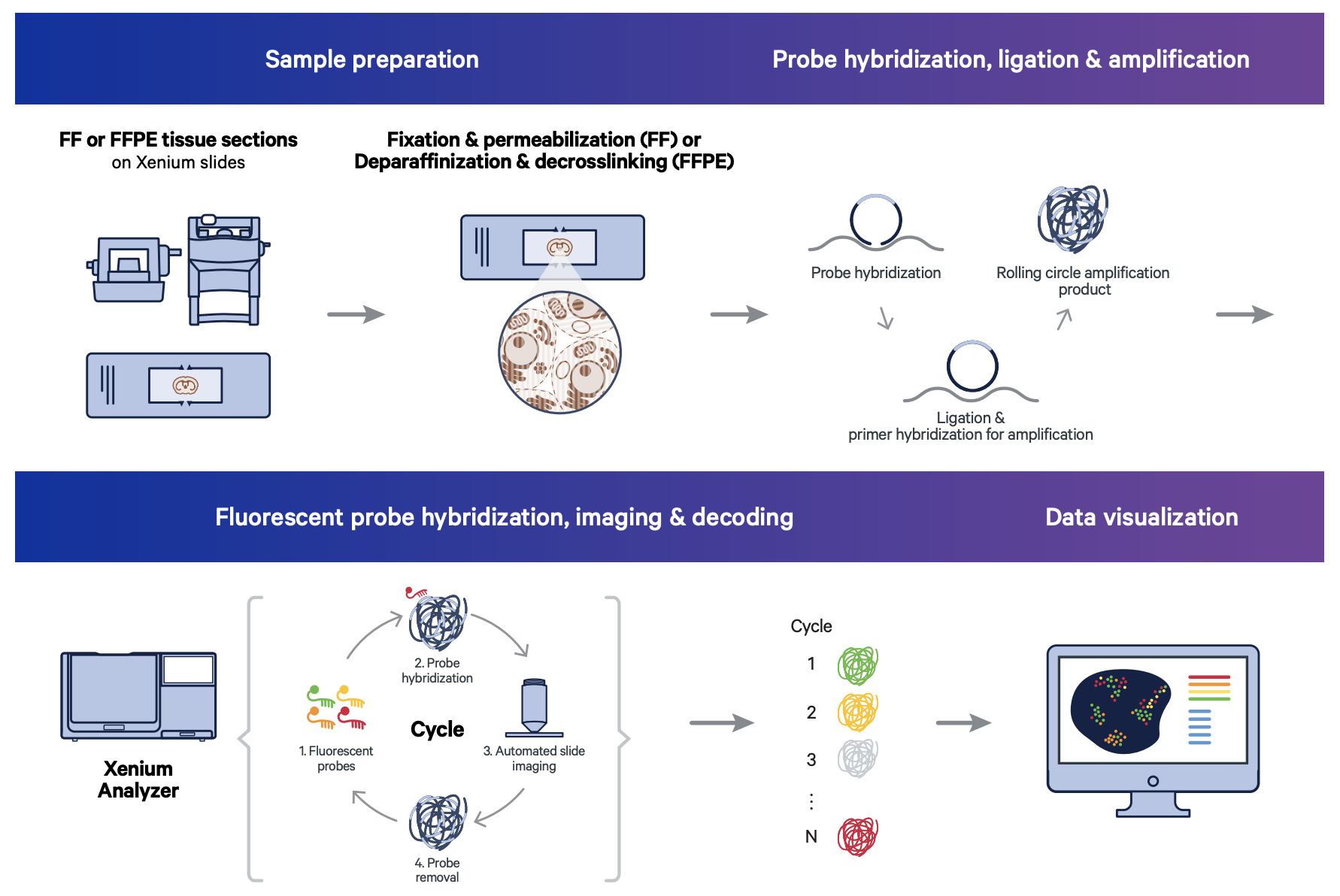
Is the assay performance the same for fresh frozen and FFPE tissue sections?
We've done pretty extensive testing on this. The sample prep protocol for fresh frozen and FFPE tissues were designed to normalize the performance as much as possible to each other. When we set up a direct head-to-head between fresh samples that have been either processed as fresh frozen or FFPE, we find that the performance is quite similar.
Of course, out in the field due to differences in histological sources and methods, there is not always a direct comparability between a fresh frozen sample and an FFPE sample. However, we have found that when sample prep is very controlled, we get very similar performance.
How thick can your sample be?
We strongly recommend that tissue blocks are sectioned at a 5-µm thickness for FFPE samples and 10-µm thickness for fresh frozen samples.
Do you design the protein probes or just RNA?
At this time, our probes are only designed for RNA transcripts. We are, however, working on launching a protein capability next year, so stay tuned to more details.
Can we get absolute gene expression or only differential gene expression of a given gene across the tissues?
You can get absolute gene expression. One of the outputs of the Xenium run is a cell expression matrix, a matrix with cells on one axis and panel genes on the other.
What is the greatest determining factor of a successful Xenium run?
Sample quality! We have a series of knowledge-based articles that give guidance on how to generate high quality blocks. It's really about controlling the dimensions of your sample going into your fixative, the temperature, minimizing the amount of time between sample collection and preservation, and the storage conditions. If you do that well, the rest of your assay off in a much better position.
Are there limitations when using Xenium to analyze highly pigmented tissues/cell types?
Generally, we have found that autofluorescence is well mitigated during the Xenium assay’s quenching step, and our analysis is able to tolerate a fairly high amount of autofluorescence typically present in a random sampling of all of our validated human/mouse tissues. Exceptionally high cases of autofluorescence tend to be associated with longer-than-optimal fixation, but we think you’d still be able to get the transcriptomic information you need, albeit at lower sensitivity.
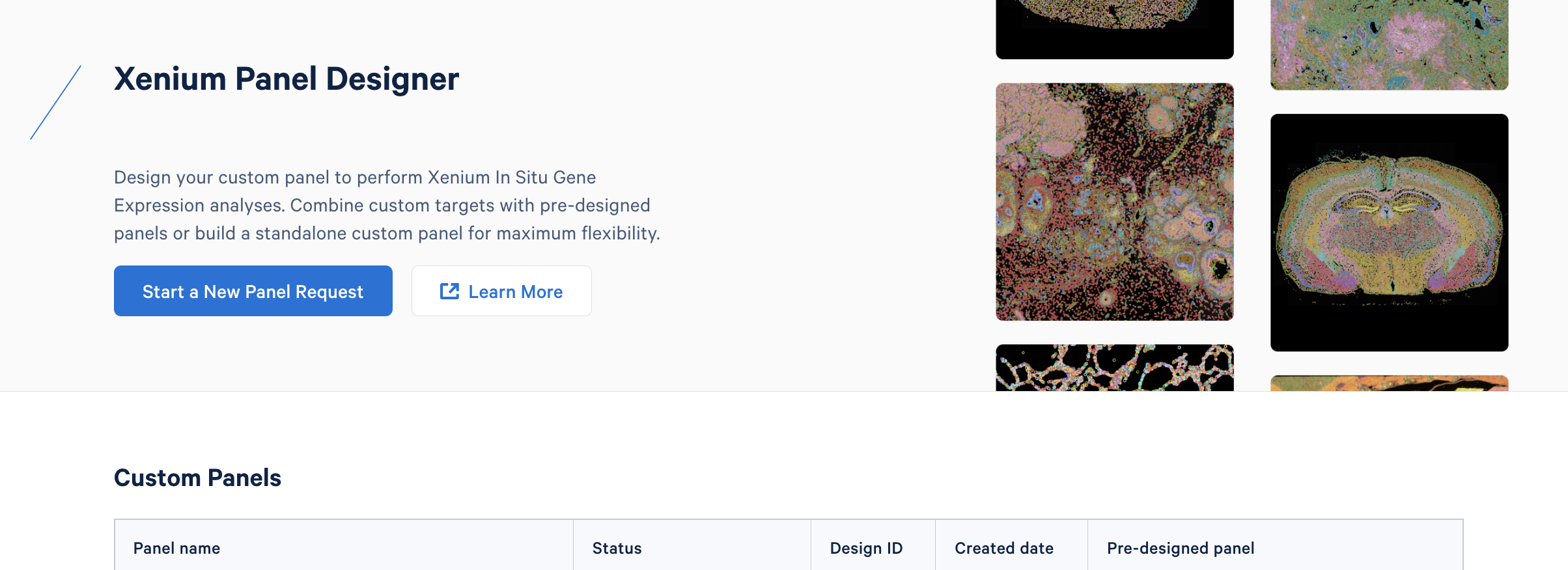
Choosing between panel options
How do I decide if I should do an add-on or fully custom panel?
It really depends on your goals of interest, and how much you think you can make use of our off-the-shelf predesigned panels. We intended the multi-tissue panels to be broadly applicable so that it would be very obtainable for most experimental systems to add on only 100 targets and still get the information you want.
We tend to find people want to go fully custom when they want to move away from cell type into more advanced applications. For example, researchers interested in looking at cell-cell interactions or pathways. In that scenario, when you are going beyond the cell typing focus of our pre-designed panels, you might need to have more than the 100 extra gene targets that the Add-on Custom Panel can accommodate.
When should I use the human skin panel over the multi-tissue panel?
If you dig carefully into the description for the Human Multi-Tissue and Cancer Panel, you'll see that skin is a supported subtype of tissue. In this case, if your question and your research project is only going to be on skin samples, we'd recommend picking the Human Skin Gene Expression Panel. There are some use cases where multi-tissue is advantageous. For example, comparing across tissues or maybe a core facility that needs to service multiple sample types.
We found internally that if you supplement a 100-gene Add-on Custom Panel to the multi-tissue panel, you can get performance very similar to one of our pre-designed panels. This is a good option to consider if you are interested if the multi-tissue panel covers a tissue that we don't have its own pre-designed panel for yet. You can get very good performance with just a bit of homework and plugging in the right genes for your Add-on Custom Panel.
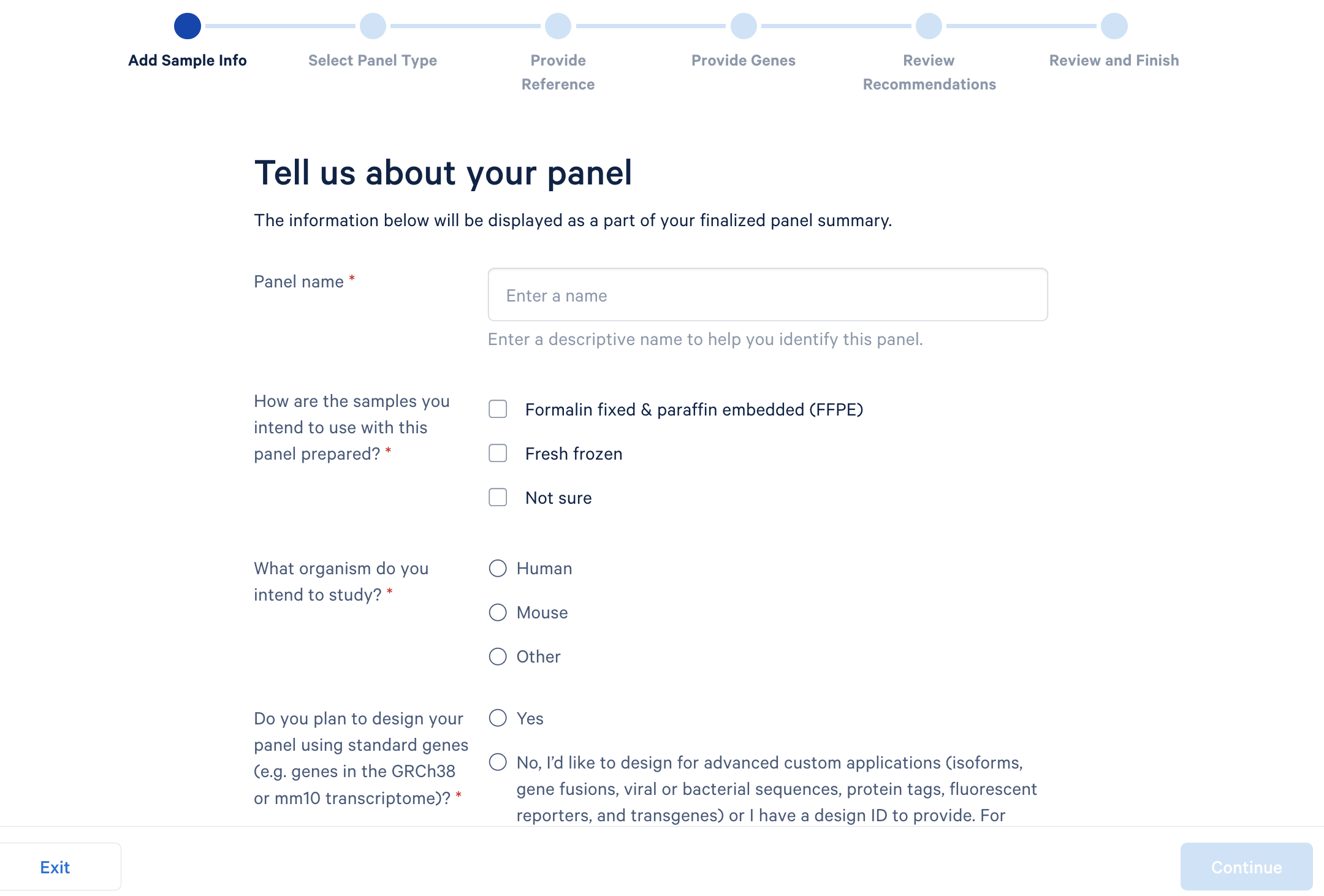
Panel and probe design considerations
What should I do if I don't have single cell reference data?
If you don't have single reference data, check the literature. You can try searching the CELLxGENE database or look for papers to find something as closely matched as possible. Totally matched is not necessarily required, but keep in mind that the further away you get between disease states, the higher the risk that the expression profile your panel is optimized against will not match what you actually see in the tissue.
If you use your own reference dataset, can you supply your own cell annotation?
Yes, so it is required to have some sort of cell annotation in order to group the cells together. Basically, you provide a text or CSV file that has a barcode and an annotated cell type for each one. At its simplest level, you can just provide the results of the graph clustering that way. But, ideally, you have some sort of more structured annotation that you provide instead.
Is there a requirement of the length of the transcript? Could you design probes to detect short sequences of around 15 nucleotides?
The minimum length that we support is 40 bases, with 20 bases for each side of the padlock. We have an advanced custom panel design tech note that goes into detail about how you can design sequences to target. If you choose to do an advanced custom upgrade, you can work with our team and we will help design those probes.
One thing to think about when you're targeting very short sequences is that they're often surrounded by constant sequences. Often you have a transcript that is long enough to target and because this technology is very specific, it is possible that you can target that 15 bases by devoting the remaining 25 bases to the adjacent constant sequence.
How can we use the panel designer for different isoforms for a given gene?
So, currently, designing against isoforms, or any targets that we consider advanced custom, is not done directly through the Xenium Panel Designer.
For isoforms and other advanced custom probe designs, you will first need to contact your sales representative to get a Design ID. Next, you will go to the Xenium Panel Designer website where you will select the option for designing against advanced targets and input your design ID. Submitting the request will get you in contact with the 10x support and design team. Our team will then work with you to design and fine-tune your custom probes.
Can you please tell me more about single nucleotide variant (SNV) target probe design?
SNVs are not officially supported at the moment for reasons that are enumerated in the advanced custom probe design tech note. As a result, depending on the sequence of your reference and mutated allele, it may or may not be amenable to this platform. However, we have had some success with SNV work as we showed at AGBT 2023. So, if you're interested in SNVs, we encourage you to reach out to your sales rep and they can help put you in contact with our advanced custom design team.
As spatial transcript data becomes more available, is there any intent to redesign standard panels around gene importance using spatial transcript data as the ground truth rather than the corresponding single cell atlas run?
That's a great question! We've been thinking about this internally and it is something we're actively looking at, so stay tuned.
This is a new field, so we try to support whatever is the latest and greatest that you, the customers, want. We're very responsive and open to what your requests are.
Want to keep learning about how to get the most out of your Xenium experiments? Here’s a couple ways to do that!
Watch the full Getting started with Xenium In Situ webinar series. Sign up for on-demand recordings →

Get more useful tips on optimizing experimental design, sample prep, and data analysis from our Experimental Planning Guide. Download now →

