Get the Most Out of Your Linked-Read Data
Linked-Reads are a powerful tool for constructing long-range information—providing a more comprehensive view of the genome and exome. But have you ever wondered how to best prepare your samples or how much sequencing you need to achieve the highest quality Linked-Read data?
We recently published a Bench Tips article with Biocompare, where we highlight important tips to make the most of our Linked-Read technology for long-range genomic analysis.
Some important items discussed are:
Sample preparation dos and don’ts.
Do:
- Pipet HMW gDNA slowly using a wide-bore pipet tip to prevent shearing.
- Elute and store HMW gDNA in Tris-EDTA buffer (not water).
- Use physical grinding instead of chemical lysis buffers when extracting gDNA from tissue samples.
Don’t:
- Use steps that can denature, nick or damage the DNA, including heat incubation, extreme pH buffers, and chemical dissociation buffers for tissue.
- Mix samples by vortexing; use brief "pulse vortexing" if absolutely necessary.
How to avoid common pitfalls in the workflow.
Our workflow is straightforward and produces barcoded, Illumina®-compatible sequencing libraries using the Chromium™ Controller with Genome or Exome reagent kits. But before you start your first experiments, it will be important to view our comprehensive users guides and demonstrated protocols located on our Support pages.
**The sequencing depth required for your application. **
We recommend >30x sequencing depth for whole genome applications, corresponding to ~800 million reads or ~128Gb for human samples. For targeted sequencing, the recommended coverage is >60x. The amount of sequencing required to achieve this depth will depend on the capture method and baits used and you can learn more about this in the Bench Tips article.
What the Loupe™ Genome Browser can show you about your Linked-Read data.
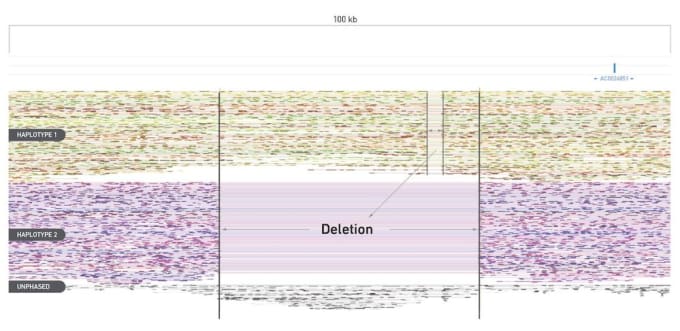
The Loupe Genome Browser provides multiple views of the Linked-Read data to aid in structural variant analysis: "Haplotypes" view, "Linked-Reads" view, and "Structural Variants" view.
The Linked-Reads view, for example, interactively and effortlessly shows you deletions by haplotype, no longer diluting your data. Here we show an approximately 30 kb deletion on Haplotype 2, with a much smaller deletion present on Haplotype 1.
Try the Loupe Genome Browser for yourself. Visit our demo site and explore pre-loaded genome and exome datasets.
We encourage you to get access to more details and more genomic information by viewing the full Bench Tips article on Linked-Reads here!
