Set the Boundaries button to view outlines for cells, nuclei, or both. Set the View cell as button to view the cell boundaries as filled, outlined, or both. These features are useful for checking cell segmentation quality (see Checking Xenium Data Quality tutorial).
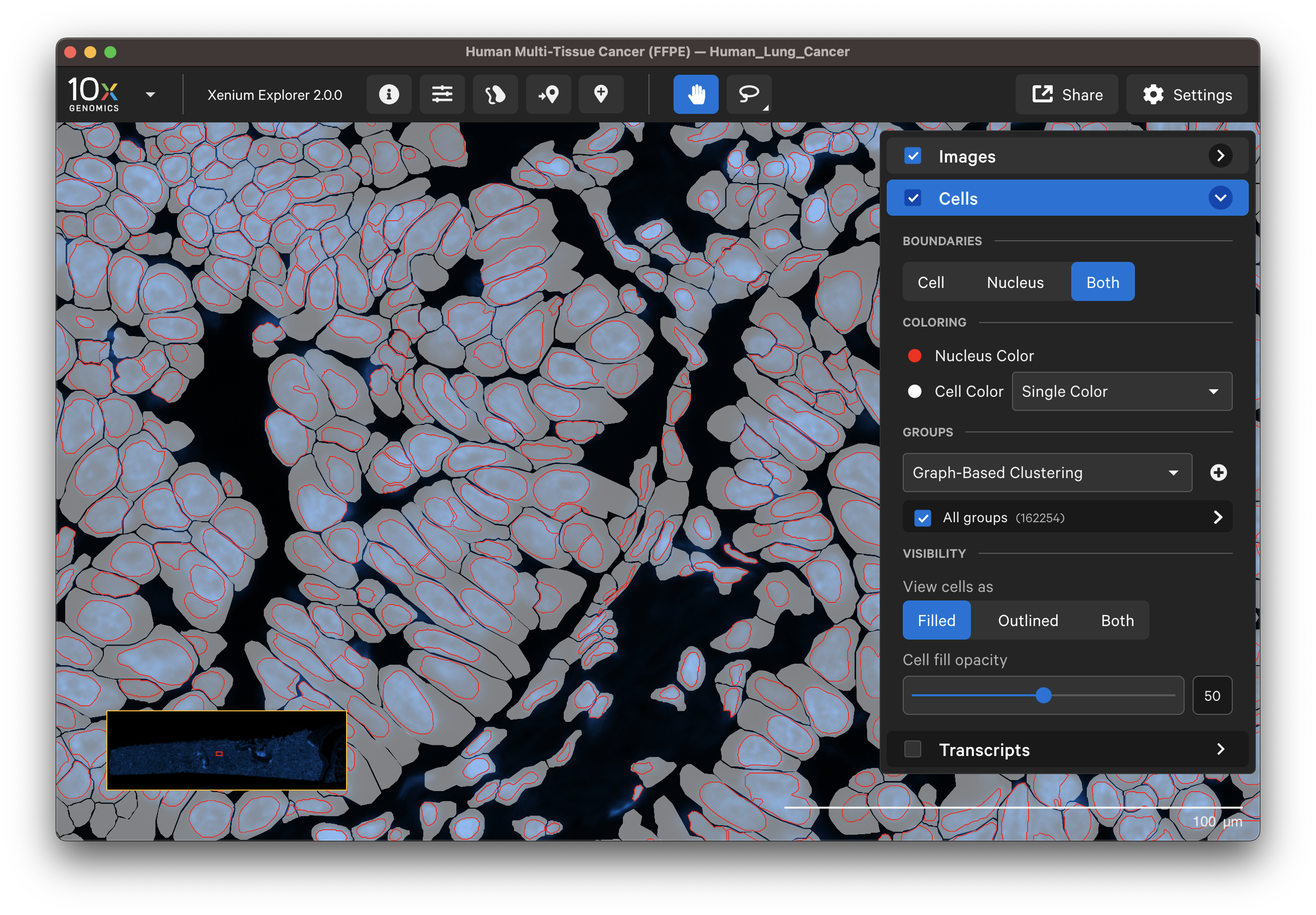
When tooltips are shown (Settings > Show Tooltips), you will see cluster, location, area, and transcript counts for individual cells as you move the cursor over each cell.
Click the link below to learn how to explore, select, and export cell statistics.
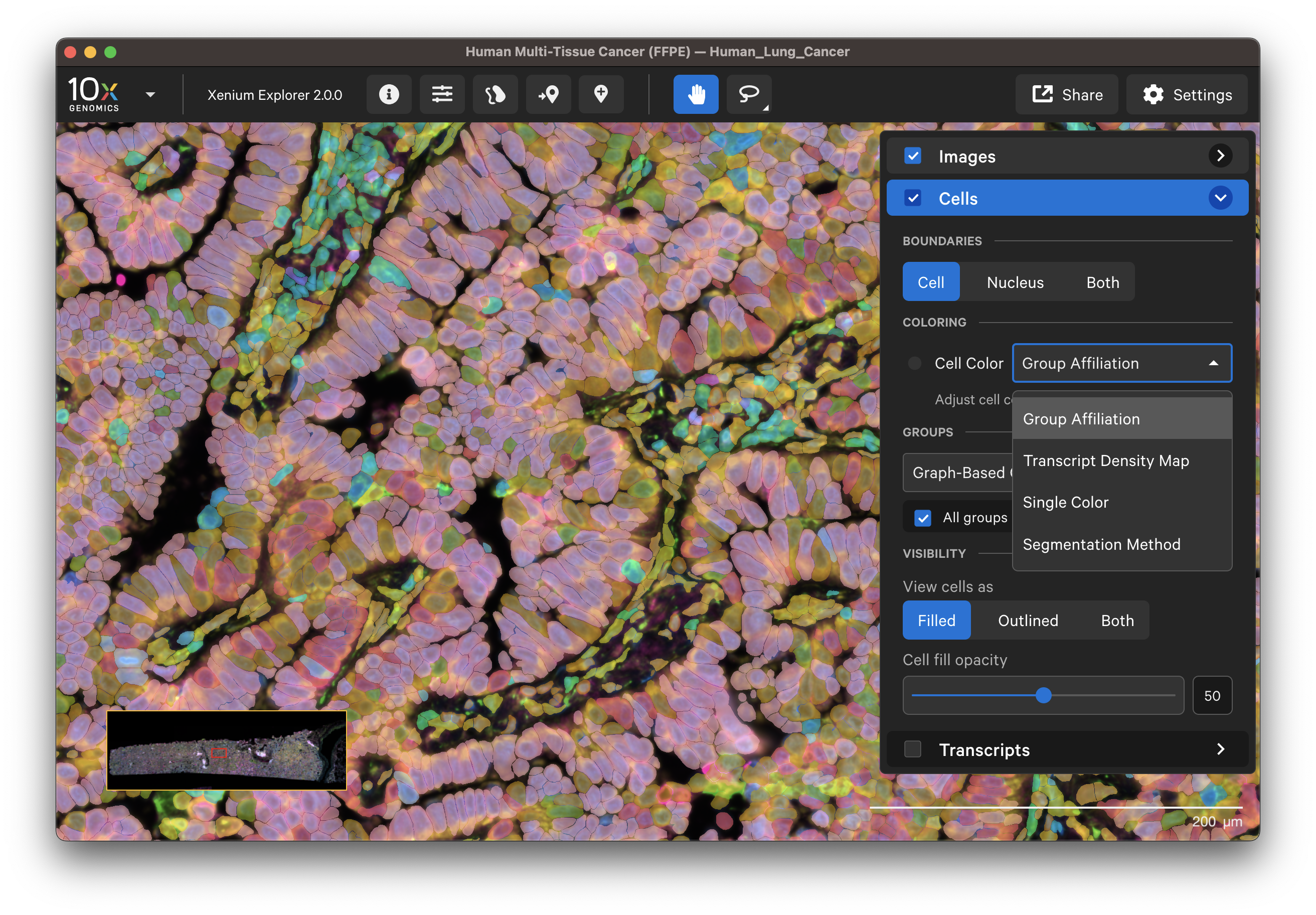
Default or custom cell colors (fill and outline) can be displayed by:
- Secondary analysis cluster group affiliation (graph-based, K-means, custom)
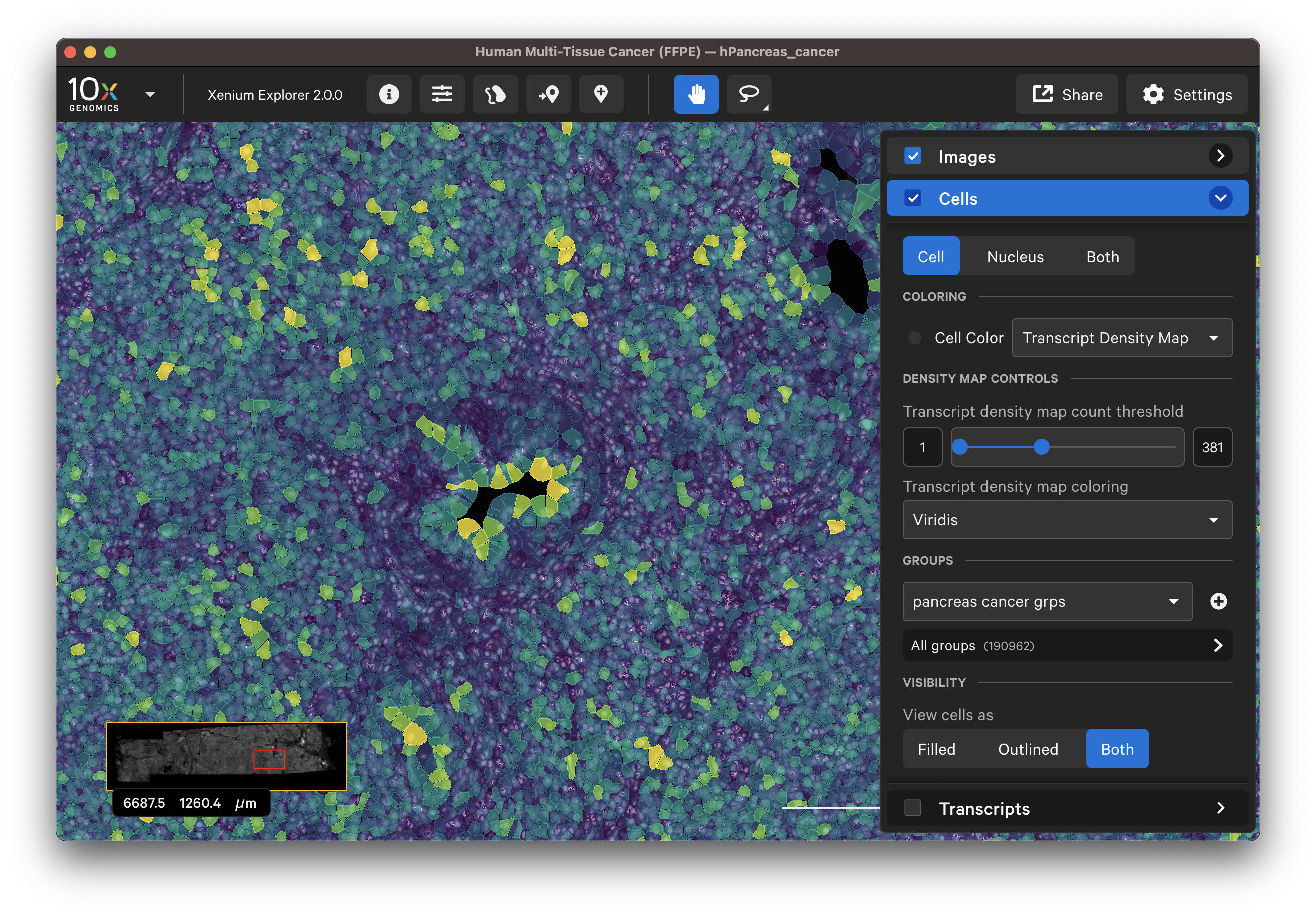
- Transcript density map
- Single color
- Cell segmentation method (for multimodal segmentation outputs only)
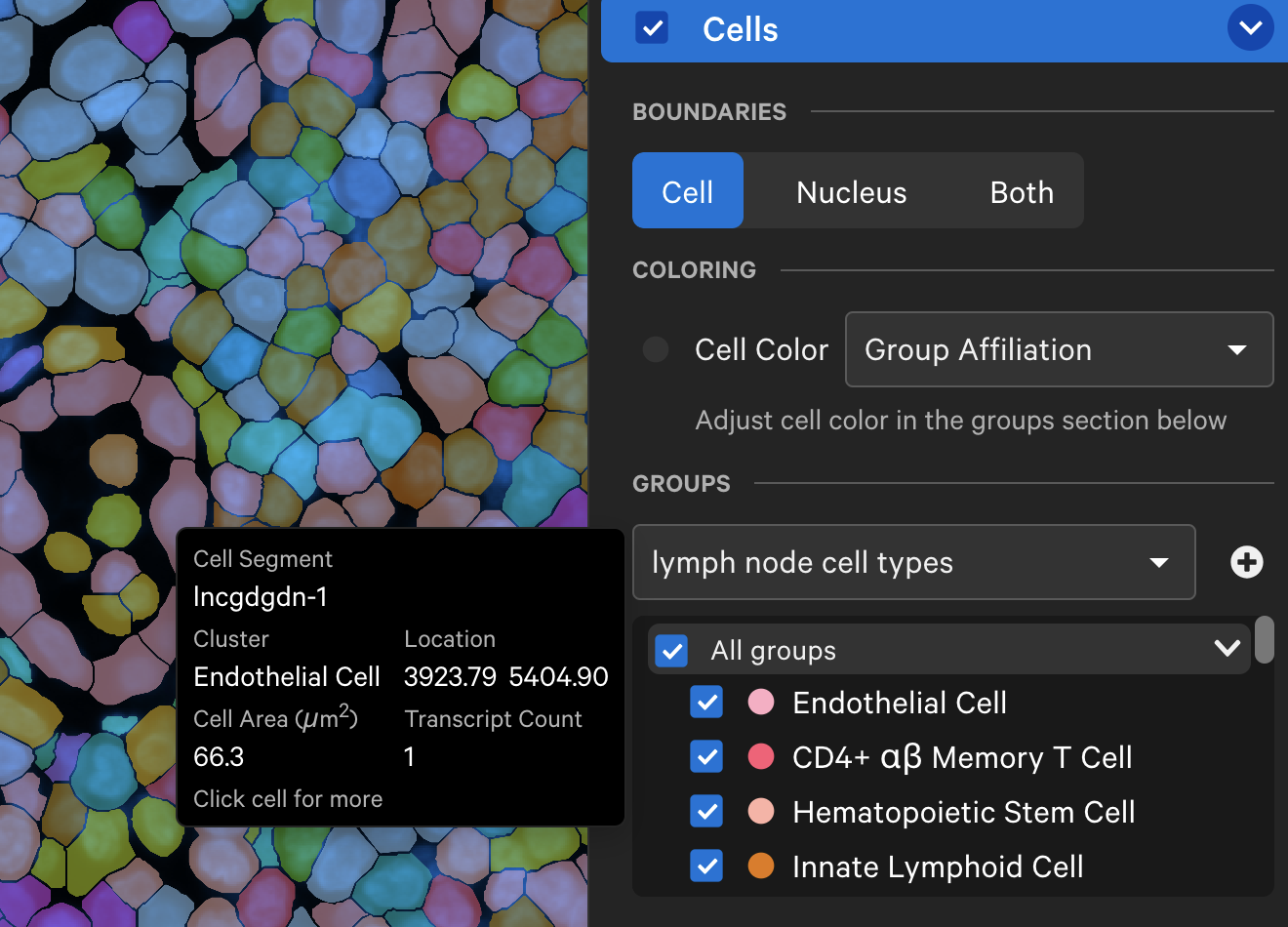
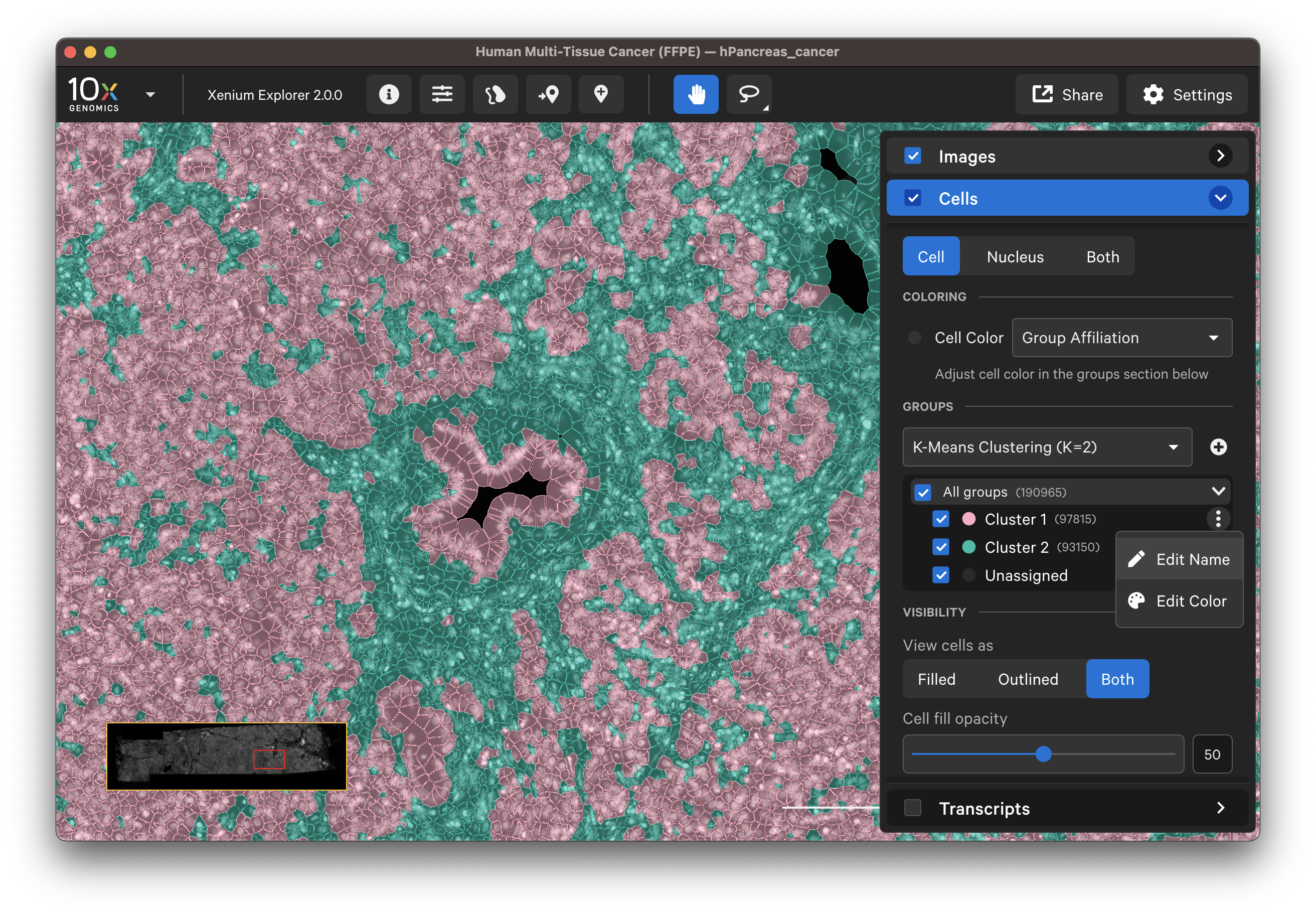
The Cells menu also shows cells colored by pre-computed graph-based or K-means cluster groups. The clusters are generated by the Xenium Onboard Analysis pipeline and are stored in the analysis.zarr.zip file. This feature is useful for assessing whether cell clusters align with known cell types or regions of interest in the sample (see Checking Xenium Data Quality tutorial).
The cluster name and color can be edited by clicking the three vertical dots next to each cluster label. You can reset the original cluster color or name by selecting the Edit Color or Edit Name and Reset to default. The clusters can be renamed based on cell type annotation using marker genes.
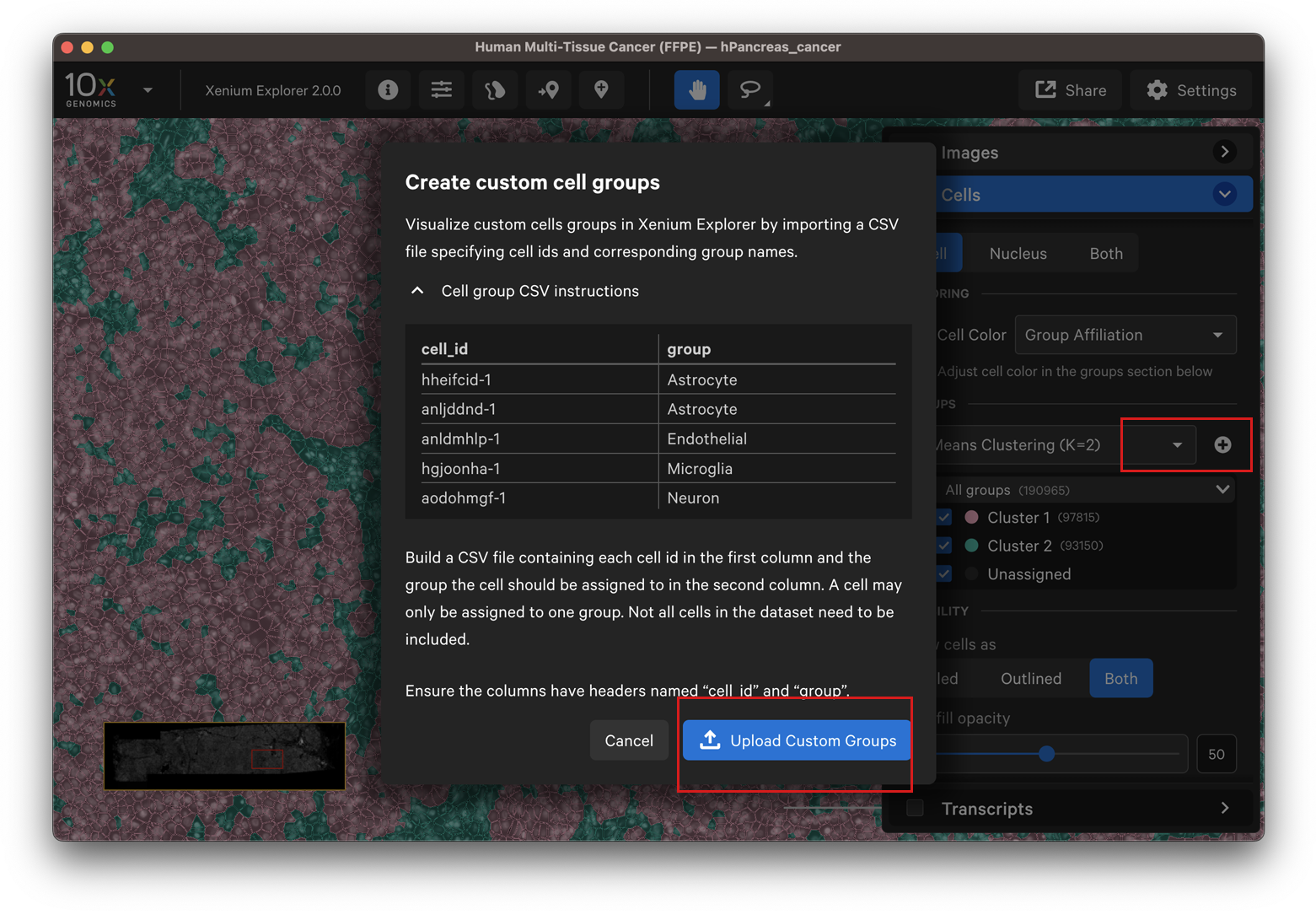
This option allows you to upload custom cell clusters as a CSV file on the Cells menu. In Xenium Explorer v3.1, custom cell groups can be uploaded without needing the precomputed analysis.zarr.zip file to be present.
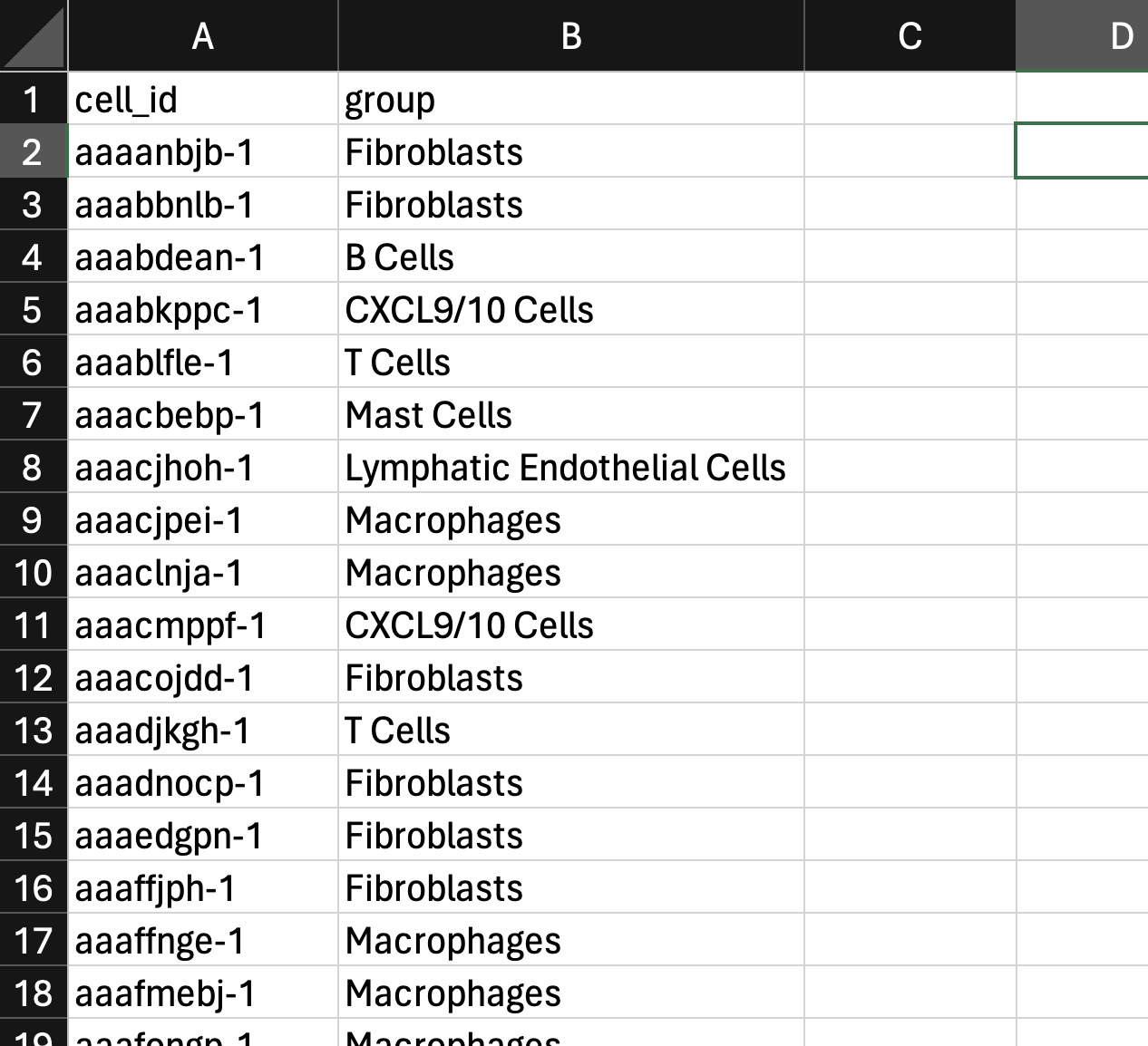
Create a cell group comma-separated value (CSV) file in a text editor, such as Microsoft Excel. The format of the CSV file is as follows:
- The first row must have two column headers in this order with these exact names: "cell_id" and "group".
- The first column is a list of cell ID names.
- The second column is a list of group labels.
- There should only be two columns in this file. At this time, multiple custom cell groupings must be uploaded as separate CSV files. You can select each group in the "Cell groups" dropdown list.
Here is an example from the pancreatic cancer dataset (download custom cell group file here):
Note that you must wait for cells to load in the window before the CSV file can be uploaded. Click the plus sign to add the CSV file. Then click Upload Custom Groups.
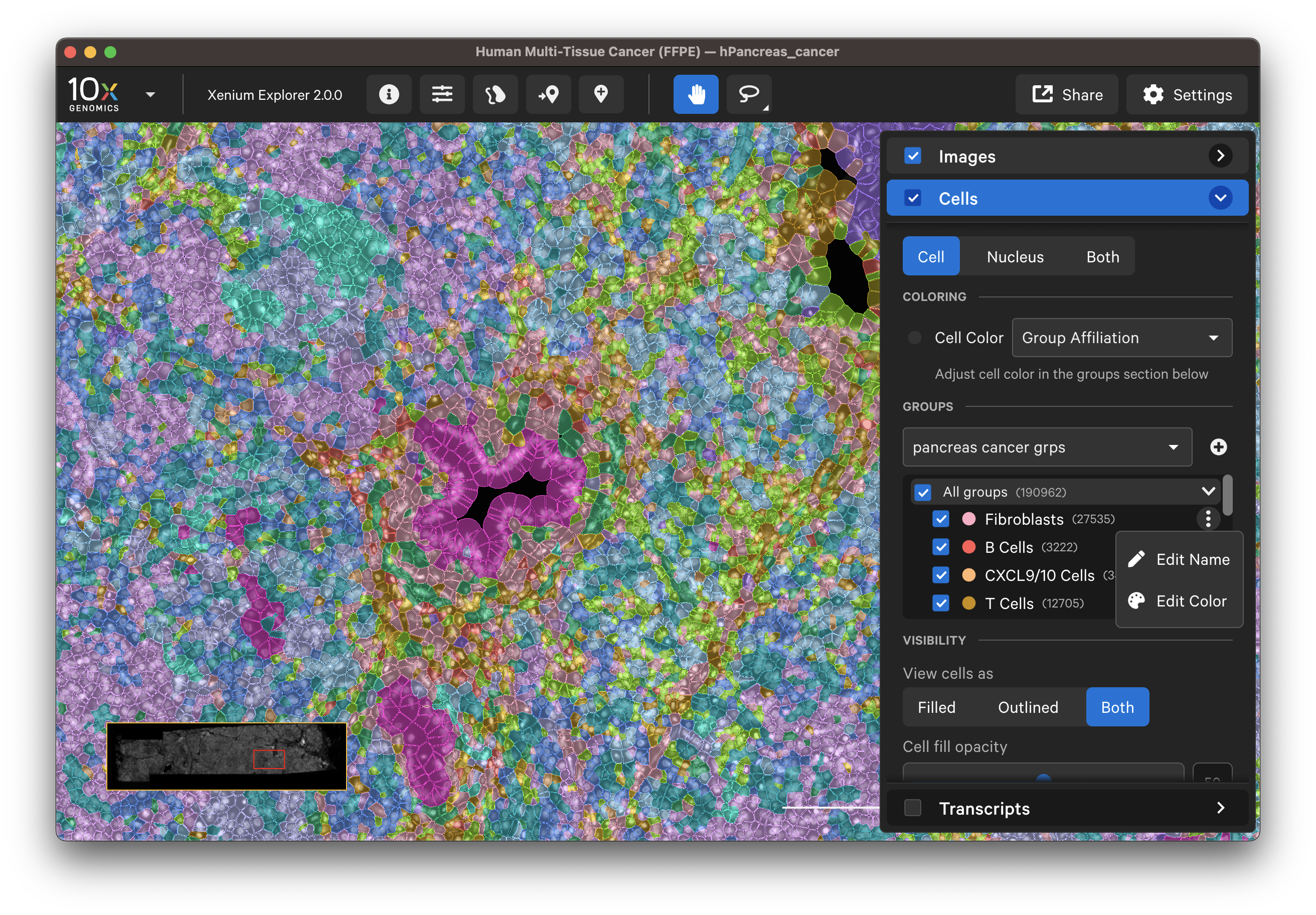
The uploaded cell groups can be saved to Saved Views. To share with colleagues, they will need to reupload the cell group CSV file. The custom cell groups look like this:
The transcript density is determined by the genes that are selected on the Transcripts menu. Cell colormap lower/upper bounds and fill opacity can be adjusted with the slider (click arrow above maximum threshold value to reset). There are several color palette options.
When colored by the transcript density map, the lowest transcript density cell maps to the lower end of the palette (i.e., viridis=purple), while the highest transcript density cell maps to the upper end of the palette (i.e., viridis=yellow), with a linear distribution in between. The Transcript density map count threshold slider allows you to choose a different mapping range, which can help to visualize cells that are at or beyond the limits of the density range.
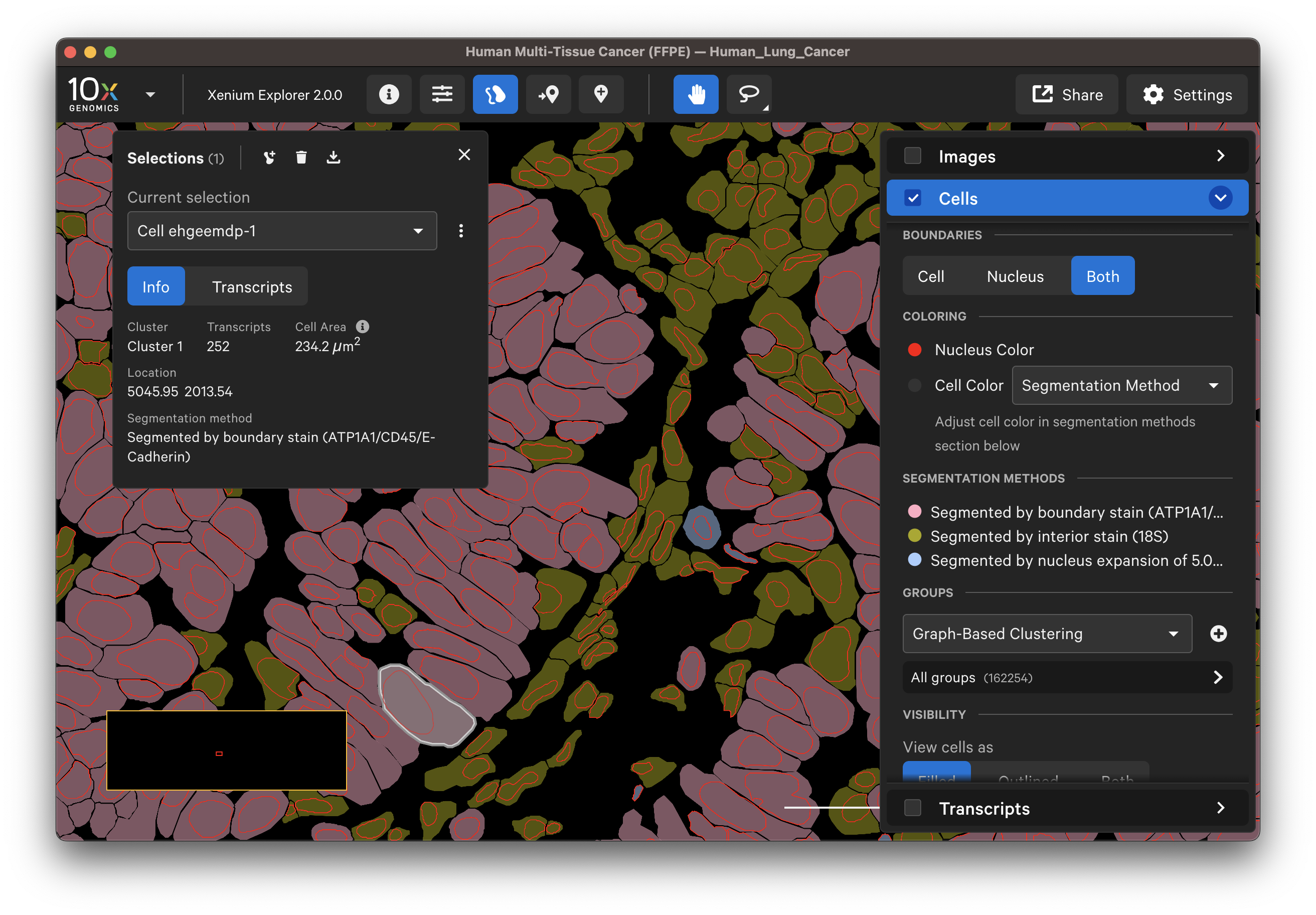
In the multimodal cell segmentation analysis workflow, three methods are used to segment cells (read more in the XOA algorithms page). You can visualize the segmentation methods in Xenium Explorer v2.0 and later. This feature is useful for qualitatively checking nucleus and cell segmentation results (see more on the checking data quality page).
To view all cells by segmentation method, select Segmentation Method in the dropdown menu for Cell Color. The inferred cell segmentation polygon boundaries are shown in Xenium Explorer. To view the segmentation method for an individual cell, click on a cell to view information in the Selection pop-up window.