The spaceranger count pipeline accepts Slide Serial Number and Capture Area arguments, in order to use the most precise fiducial and spot coordinates for an experiment. Starting with Space Ranger v2.0, it is important to also note the slide version. The following illustrations highlight the key components of a Visium slide as well as the different slide versions.
| Capture Areas | Slide Serial Number |
|---|---|
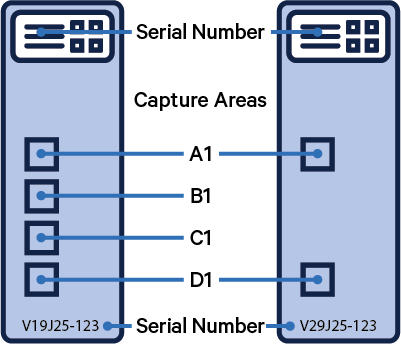 |  |
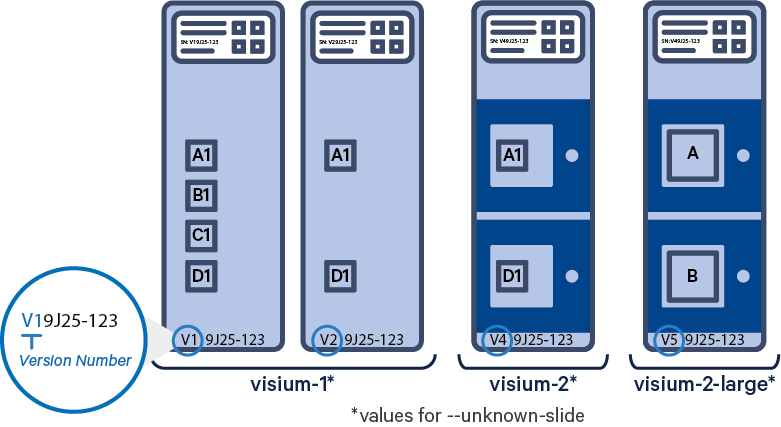
This table lists the slide version, sample type and workflow compatibility:
| Sample Type | Supported Slide Versions | Capture Areas | Supported Workflows |
|---|---|---|---|
| Fresh Frozen (FF) | V1 (e.g. V19L01-041) | 4 | Spatial Gene Expression |
| V2 (e.g. V20S03-001) | 2 | Spatial Gene Expression | |
| Formalin-fixed Paraffin-embedded (FFPE) | V1 (e.g. V19L01-041) | 4 | Spatial Gene Expression |
| V2 (e.g. V20S03-001) | 2 | Spatial Gene Expression | |
| V4 (e.g. V49J01-123) | 2 | Spatial Gene Expression CytAssist Compatible | |
| V5 (e.g. V59J01-123) | 2 | Spatial Gene Expression CytAssist Compatible |
CytAssist also supports Fresh Frozen (FF) and Fixed Frozen (FxF) sample types on V4 and V5 slides using probes against whole transcriptome.
There are three different ways to specify the slide information in spaceranger count run.
| Space Ranger Arguments | Slide Specific Layout File | Pipeline Internet Access | Details |
|---|---|---|---|
--slide --area | Yes | Yes | The pipeline will download the layout file associated with the supplied slide serial number. |
--slidefile --slide --area | Yes | No | User downloads a slide layout file for a Visium slide before running the pipeline. Download here |
--unknown-slide (one of --visium-1, --visium-2, or --visium-2-large) | Yes | No | The pipeline uses a default slide layout file for spot and fiducial coordinates. The typical per-spot difference between the default layout and a specific slide is under 10 microns. |