More Complete HLA Analysis
The Major Histocompatibility Complex (MHC), or the human leukocyte antigen (HLA) locus, is a highly diverse region of the genome containing genes involved in regulation of the immune system. It has been associated with diseases, such as autoimmune disorders and complement deficiency, and is critical in organ transplantation, where the HLA alleles of the donor must match those of the recipient to avoid graft vs. host disease.
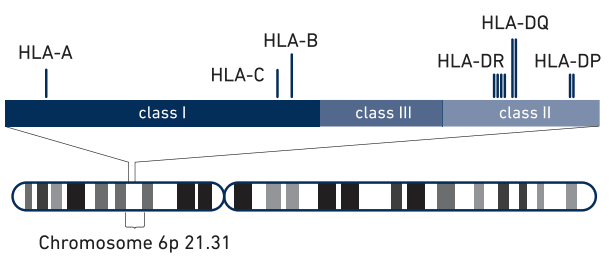
The schematic(1) above shows the class I, class II and class III regions, with the HLA class I and class II genes highlighted. Haplotype reconstruction across these genes is critical for HLA type matching.The MHC region is more diverse than the rest of the genome and contains not only single nucleotide variations (SNVs), but also large-scale structural variation such as copy number variations (CNVs) and structural variants (SVs) such as inversions. As a result, this region is quite difficult to analyze using standard short read next generation sequencing (NGS), requiring the development of several specialized assays and techniques.
Linked-Read data generated by the Chromium™ Genome Solution along with Long Ranger™ Software enables HLA analysis using short read NGS instruments. The 10x barcodes added during library preparation allow short reads originating from the same HMW DNA molecule to be bioinformatically linked back together, creating Linked-Reads. When applied to the MHC, Linked-Reads are used to both identify variants and reconstruct each allele faithfully over the length of the HLA gene region.
In the new application note, "Chromium™ Genome Solution and Long Ranger™ for HLA Analysis", we demonstrate the ability to completely phase all of the classical HLA genes, as well as the class 2 HLA genes, with only a single break in the HLA phase block. Read more here: /resources/application-notes
- Xie et al., 2010 BMC Bioinformatics doi:10.1186/1471-2105-11-S11-S10
