Discovering Rare Cell Types with Single Cell RNA-Seq
Guest Author: Jennifer MacArthur
The Human Cell Atlas initiative seeks to identify and map every cell type in the human body, laying the foundation for high-resolution biology. These atlasing approaches are deepening our understanding of cell types and subtypes, identifying cells believed to be previously unknown, and 10x Genomics is facilitating this kind of research, building products that can characterize cells at fundamental resolution and massive scale. Our Single Cell Gene Expression Solution has helped researchers find rare and unexpected cell types in many kinds of tissues, including the lung, kidney, colon, and liver. Here, we highlight some of the discoveries that are reshaping our understanding of fundamental biology and advancing disease insights.
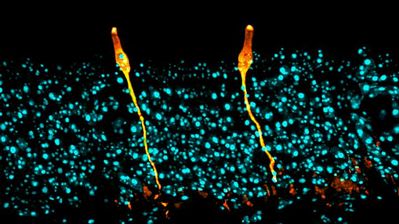
New lung cell type advances our understanding of cystic fibrosis
The discovery of the "pulmonary ionocyte" demonstrates how identifying new cell types can be immediately useful to disease research. When a team at Massachusetts General Hospital and the Broad Institute used single cell gene expression to catalogue cells that line the airway, they found seven cell types, instead of the expected six. The novel cell population is similar to salt-balancing ionocytes in fish gills or frog skin. Although they represent only 1% of airway epithelial cells, pulmonary ionocytes are the primary source of CFTR gene activity. This discovery redirects efforts to correct CFTR mutations, as cystic fibrosis researchers have previously focused gene therapy approaches on ciliated cells (1).
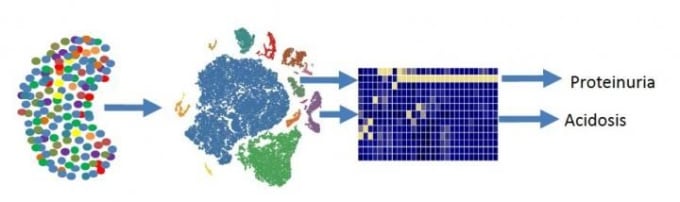
Unexpected kidney cell type may contribute to metabolic acidosis
The kidney is a sophisticated organ with specialized cell types to support complex physiological tasks, like balancing electrolytes and regulating blood pressure. Researchers at the University of Pennsylvania used single cell gene expression to make a comprehensive map of the mouse kidney. From nearly 58,000 cells, they identified 18 previously described kidney cell types, along with three novel cell populations. One new cell type was a group of transitional cells in the collecting duct. This showed that cells focused on sodium/potassium balance can switch, becoming cells focused on acid/base balance (and vice versa). They also mapped kidney disease-associated genes to specific cell types to help infer cellular drivers of diseases, such as proteinuria and metabolic acidosis (2).
New cells in the gut could be a target for inflammatory bowel disease treatment
Single cell gene expression profiling is also letting researchers compare healthy and diseased tissues with fundamental detail. Scientists at Oxford University analyzed over 11,000 cells from the gut lining of healthy volunteers and patients with ulcerative colitis. This revealed a gene expression gradient and differentiation hierarchy that ascends through the crypt, and helped dissect cell-type-specific responses to inflammation. The ten cell types identified in healthy colon epithelium included a novel pH-sensing absorptive colonocyte, which is lost during inflammation. Two additional cell types were observed in inflamed tissue: goblet cells that express a major determinant of barrier maintenance, and intraepithelial immune cells. These insights offer potential new targets for inflammatory bowel disease treatment (3).
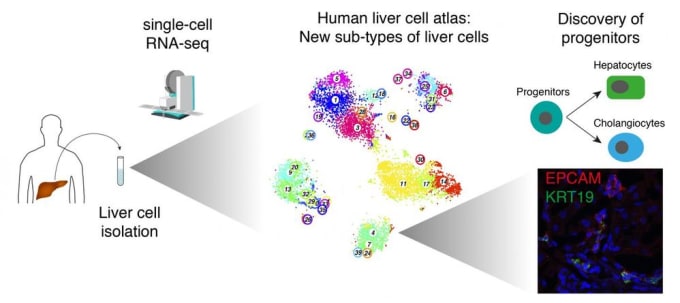
Liver cell atlas reveals new precursor cells related to liver regeneration
This summer, a team at Max Planck Institute published a detailed map of cell populations in the human liver. They performed single cell gene expression profiling of over 10,000 liver cells from nine healthy donors. This high-resolution analysis revealed subtypes of endothelial cells, Kupffer cells, and hepatocytes with discrete gene expression profiles, which are believed to be previously unknown. They also identified a rare subpopulation of bile duct cells capable of forming organoids. This precursor cell type can differentiate into either hepatocytes or bile duct cells, and may play an important role in liver regeneration (4).
As our customers all over the world are demonstrating, high-throughput single cell RNA sequencing is a powerful discovery tool for rare cell types. Even in systems that have been studied for decades, there are cells we hadn’t noticed before—their signal hidden under bulk RNA-seq data or not discernible by preselected markers. Our solutions offer a combination of comprehensive genomics and single cell resolution that is shedding new light on complex biology and disease mechanisms.
- D. Montoro, A.Haber, M. Biton et al., A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature. 560, 7718 (2018).
- J. Park, R. Shrestha, C. Qiu et al., Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science. 360, 6390 (2018).
- K. Parikh, A. Antanaviciute, D. Fawkner-Corbett et al., Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature. 567, 7746 (2019).
- N. Aizarani, A .Saviano, Sagar et al., A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. 572,7768 (2019).
