Direct fixation of whole blood samples for simplified single cell profiling
Since the writing of this blog, we have introduced GEM-X Flex Gene Expression which has lower input requirements and additional performance gains. Click here to learn more.
Blood is often collected during routine clinical procedures and over the course of research projects, such as clinical trials or longitudinal studies. Clinicians and researchers can access a wealth of information from blood, including insights into disease progression and drug efficacies and toxicities. But due to rapid degradation of biomolecules—including RNA—analyses are often limited to protocols that can be performed within 24 hours of collection.
We developed a protocol for direct fixation and storage of whole blood that preserves RNA integrity and single cell information to overcome this limitation. This easy and robust sample prep workflow enables reliable, sensitive scRNA-seq of these important but tricky samples with the Chromium Single Cell Gene Expression Flex assay.
Read on to learn more about:
- Challenges and benefits to single cell analysis of blood samples
- Key advantages of our first-of-its-kind protocol
- Advice from 10x Genomics’ expert scientists
The complex challenges to single cell analysis of blood samples
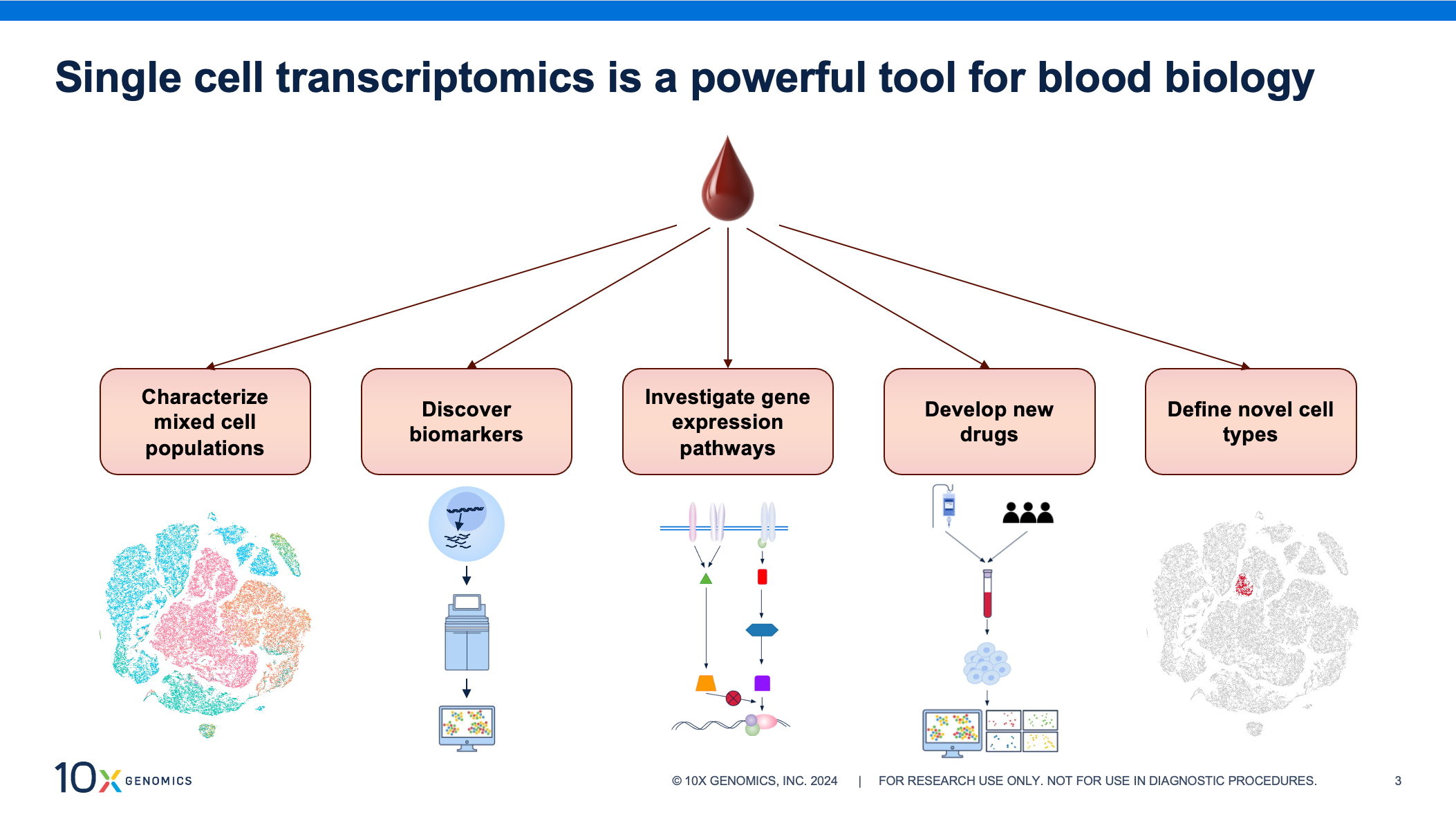
Blood samples are a convenient source of biological information as collection is non-invasive and routine, making samples readily available. Transcriptomic profiling of these samples at single cell resolution can provide invaluable insights, aiding in identification of prognostic biomarkers, investigations into mechanisms of disease, and identifying novel drug targets in fields like autoimmunity, oncology, and infectious disease.
But moving quickly from blood collection to cell isolation during sample processing is critical for capturing accurate biology. Researchers have noted that a time delay at this stage can lead to changes in gene expression profiles and negatively impact downstream results (1, 2, 3), potentially hindering groundbreaking insights. The effect can be even greater when aiming to generate large datasets, conduct longitudinal studies, or compare data collected at multiple institutions. Rapid processing might not always be feasible, but consistency in sample handling across experiments is key to limiting technical artifacts, retaining accurate biology, generating high-quality data, and avoiding skewed analysis.
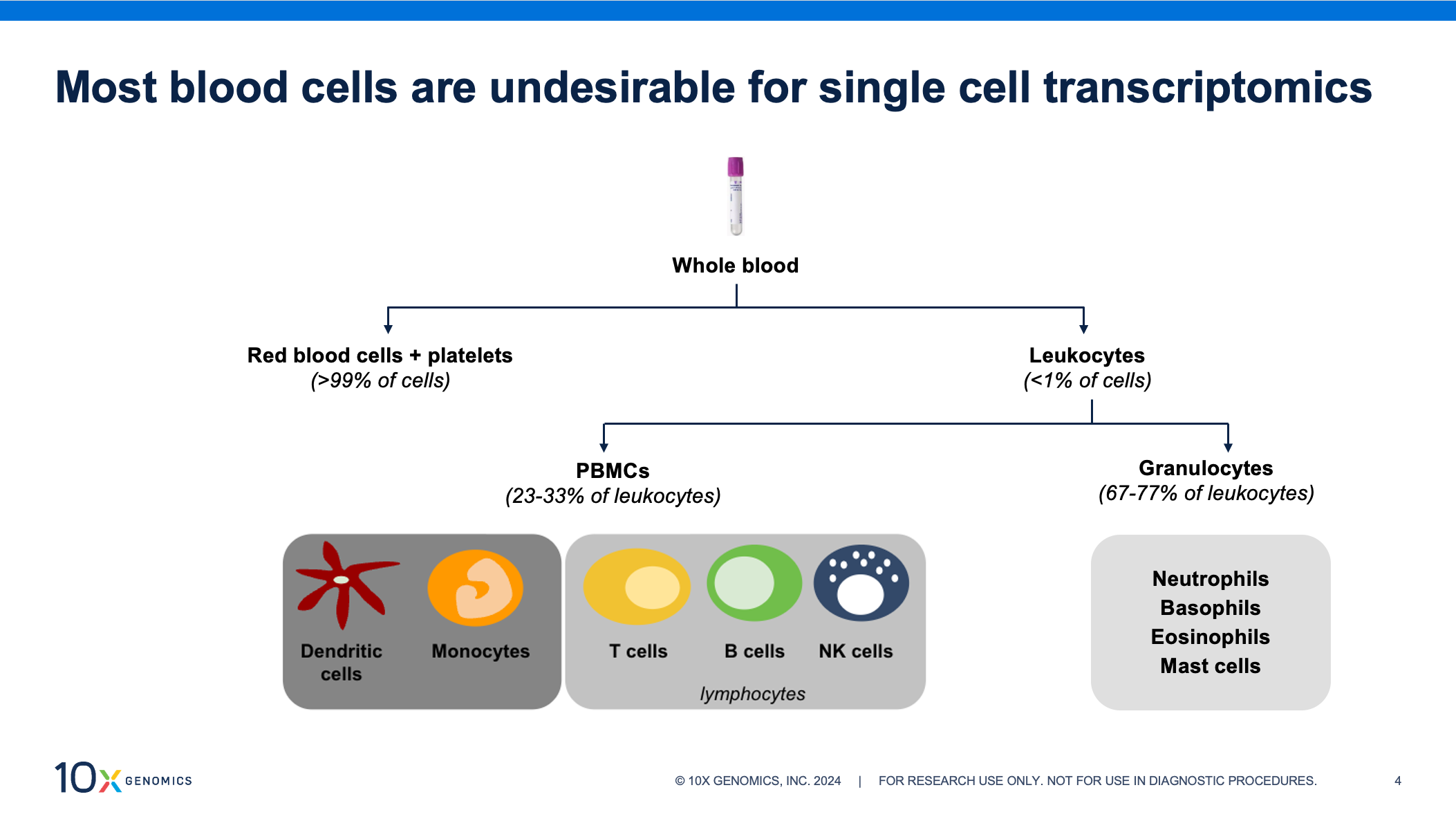
White blood cells, or leukocytes, make up less than 1% of blood cells yet contain nearly all of the transcriptomic information and should be enriched from whole blood prior to single cell assays. Leukocytes are a mixed cell population of peripheral blood mononuclear cells (PBMCs) and granulocytes—fractions with different stabilities and handling requirements. PBMCs are composed of dendritic cells, monocytes, and lymphocytes (T cells, B cells, and natural killer [NK] cells). Of the two, this is the more stable fraction. Refrigeration is necessary to prevent transcriptomic changes, but they can be enriched from refrigerated blood for up to 48 hours after collection. Granulocytes account for over half of circulating white blood cells and are composed of neutrophils, basophils, eosinophils, and mast cells. This group can deteriorate quickly if refrigerated, and we recommend enriching within two hours of collection, if possible. Though more prevalent than PBMCs, granulocytes remain understudied and are often excluded from clinical trials, in part because of their fragility.
If researchers are going to investigate these samples at single cell resolution, maintaining accurate biology is the first step to revealing actionable results. For this to be possible, reliable and consistent processing is a must.
So what are the rewards for careful handling and analysis? A wealth of information.
Guo et al. used scRNA-seq to explore how aging influences changes in populations of NK cells—key players in antitumor and antiviral responses. They identified a unique memory-like NK subset that accumulated with age and demonstrated cell functionality positively correlated with more severe cases of COVID-19 (4). Recent work from Ng et al. used single cell profiling to provide insights into how neutrophils can be reprogrammed by the tumor microenvironment. Their team highlighted how this adaptability to environmental cues could lead to new immunotherapy targets, pursuing specific neutrophil responses (5).
And these two examples are just scratching the surface.
So we have two incredibly important cell fractions at odds with each other’s storage conditions and with the logistical concerns around processing for single cell analysis. If we look to blood preservation techniques that limit RNA degradation, we find methods that have demonstrated compatibility with downstream bulk RNA sequencing, but the same has not been true for single cell assays. Retaining intact cells during blood preservation has proven to be a challenging obstacle.
What if all of these conditions weren’t standing in your way? What if you could introduce stability to your samples while preserving all of these cell types in such a way that you could easily perform high-quality single cell RNA-seq? Our team has made that possible—enabling you to apply simple, standardized fixation steps to whole blood samples, avoid the urgency of cell enrichment and isolation, and increase confidence in your results.
Back to topA flexible solution for single cell analysis of clinical samples
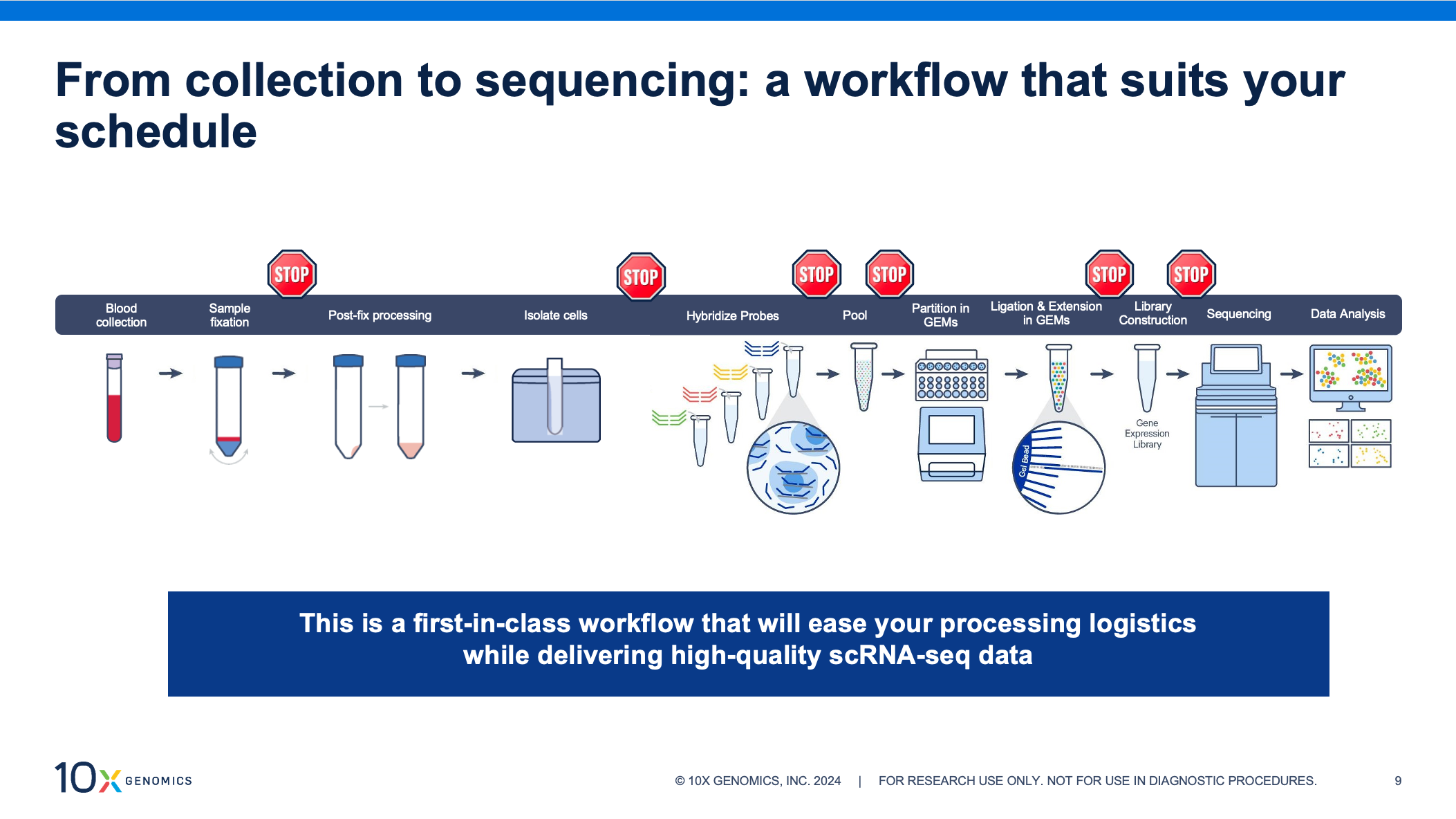
Our new sample prep workflow lets you perform scRNA-seq on blood samples at your own pace. This protocol is the first of its kind, enabling direct fixation of whole blood for downstream analysis with our Chromium Single Cell Gene Expression Flex kit. Now you can store and transport these critical samples, and isolate cells on a flexible schedule or at a central location.
Sample fixation is integral to the Gene Expression Flex workflow, making it incredibly well-suited for this new application. By fixing samples at the point of collection, you can store and transport samples without compromising data quality, preserve fragile cells, and capture transient cell populations or states. This highly sensitive, probe-based assay improves results for challenging samples, demonstrating high performance with low RNA–content cells and fragile cells that don’t hold up well under other storage conditions. Adapting this workflow to bring convenience and confidence to single cell profiling of white blood cells was a natural fit.
Along with the ability to store and transport your samples, fixation with the Flex assay lets you customize your workflow with multiple, optional stopping points. This means you can process samples in batches—even from multiple collection sites—and run them later, on your schedule. Whole blood fixation, in particular, brings two stopping points to the sample collection phase of the workflow: after fixing your whole blood samples and after cell isolation.
In a recent webinar, 10x Genomics scientists discussed the development of this new sample prep workflow and explored performance data. You can access the full webinar on demand for protocol details, tips and tricks that save time and improve performance, and analysis of comparative data from cell types under different storage conditions, including samples from healthy donors vs. those with cancer and autoimmune diseases.
Keep reading for a preview of the data, Q&A highlights, and additional resources.
Whole blood fixation made quick and easy
Now, scRNA-seq in translational research has the benefit of an optimized, user-friendly workflow for direct fixation of whole blood. The steps are quick and easy, and the robust protocol yields consistent results across users and samples. How simple is the fixation process? Let’s take a look.
First, blood is collected and added to fixation buffer. Then, samples are incubated according to your cell fraction of interest. For downstream PBMC isolation, samples should be fixed at 4°C overnight or for up to 1 week. If you will be enriching for all leukocytes (including granulocytes), samples should be fixed at room temperature overnight or at 4°C for 1 week. At this point, you can move forward with cell enrichment or choose to resuspend your sample in 50% glycerol and store at –80°C for up to 1 month.
Without fixation, it is best practice to enrich your cells of interest immediately in order to retain biological accuracy. Isolation of either PBMCs or leukocytes from an individual sample takes around an hour, which can make for a laborious schedule, depending on when samples arrive throughout the day or week. However, with whole blood fixation, hands-on time for the fixation protocol is only 5 to 15 minutes per sample.
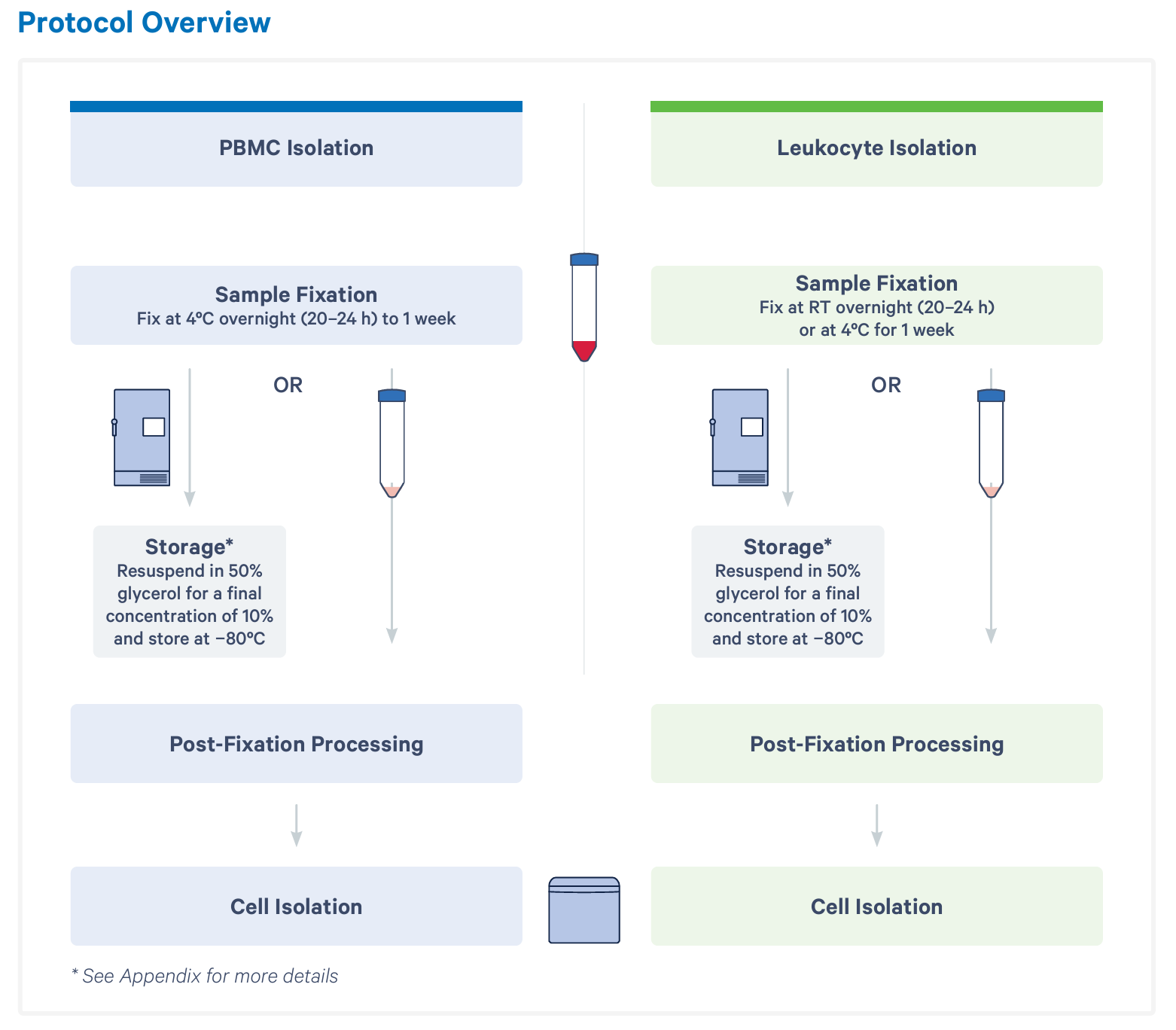
Regardless of when you choose to move forward with your fixed samples, your next steps are post-fixation processing and cell isolation. Together, these steps take 1 to 2 hours, depending on how many samples you’re working with at once. A single user can comfortably handle 8 at a time. Here, you have an optional stopping point before moving on to probe hybridization and the rest of the scRNA-seq workflow.
Preserve critical biology and capture fragile cell types for high-quality scRNA-seq
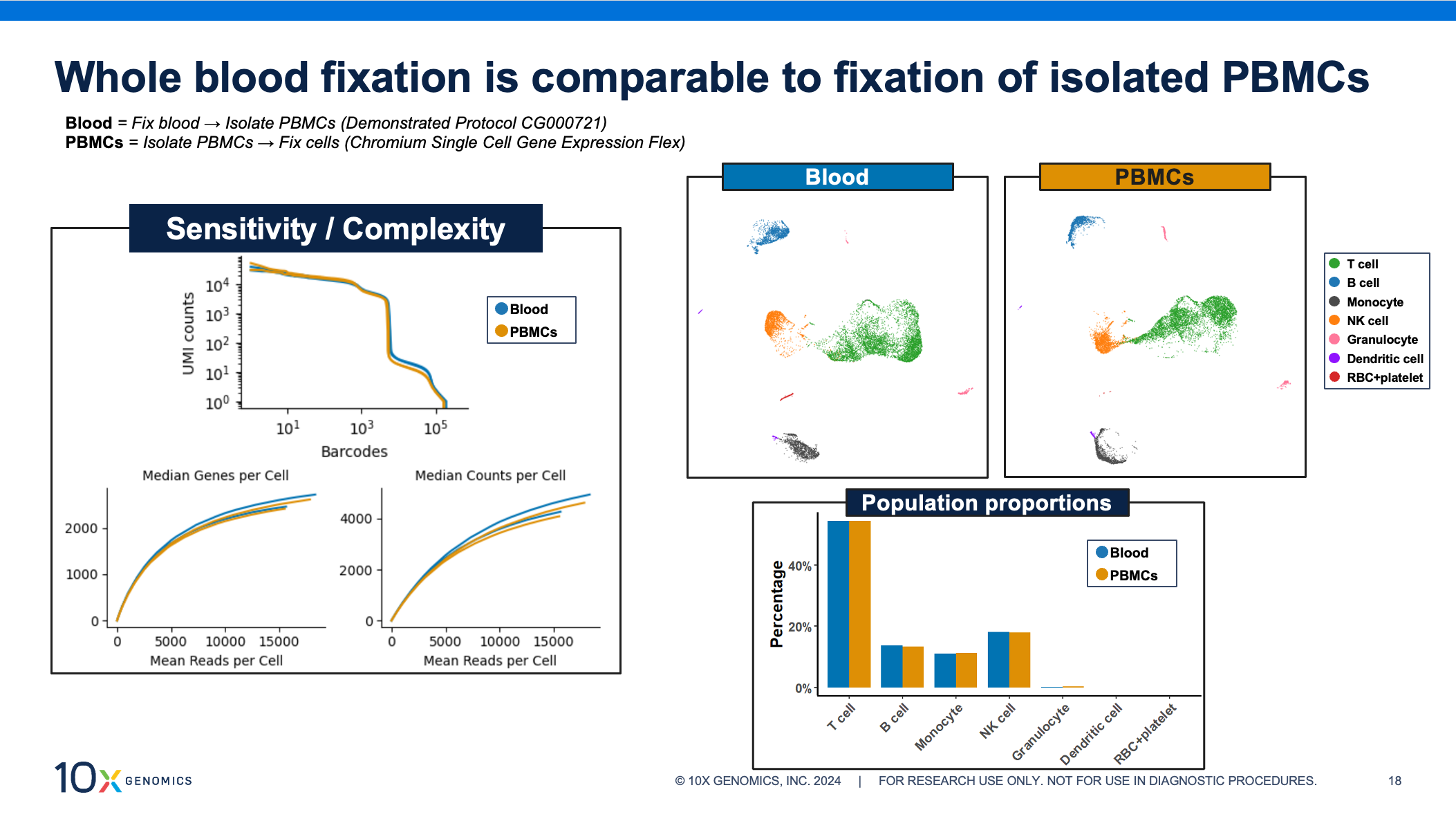
All of the fixation, processing, and cell isolation steps have been optimized to yield high-quality scRNA-seq data results from whole blood samples. Gene Expression Flex shows excellent results when fixing purified single cell suspensions, and we expect the same for these fixed isolated cells. In the figure below, we assess data for PBMCs, comparing both sample processing workflows side by side. We observe that whole blood fixation provides extremely similar complexity and sensitivity to isolated PBMCs. Additionally, both approaches show similar cell clustering and nearly identical population proportions.
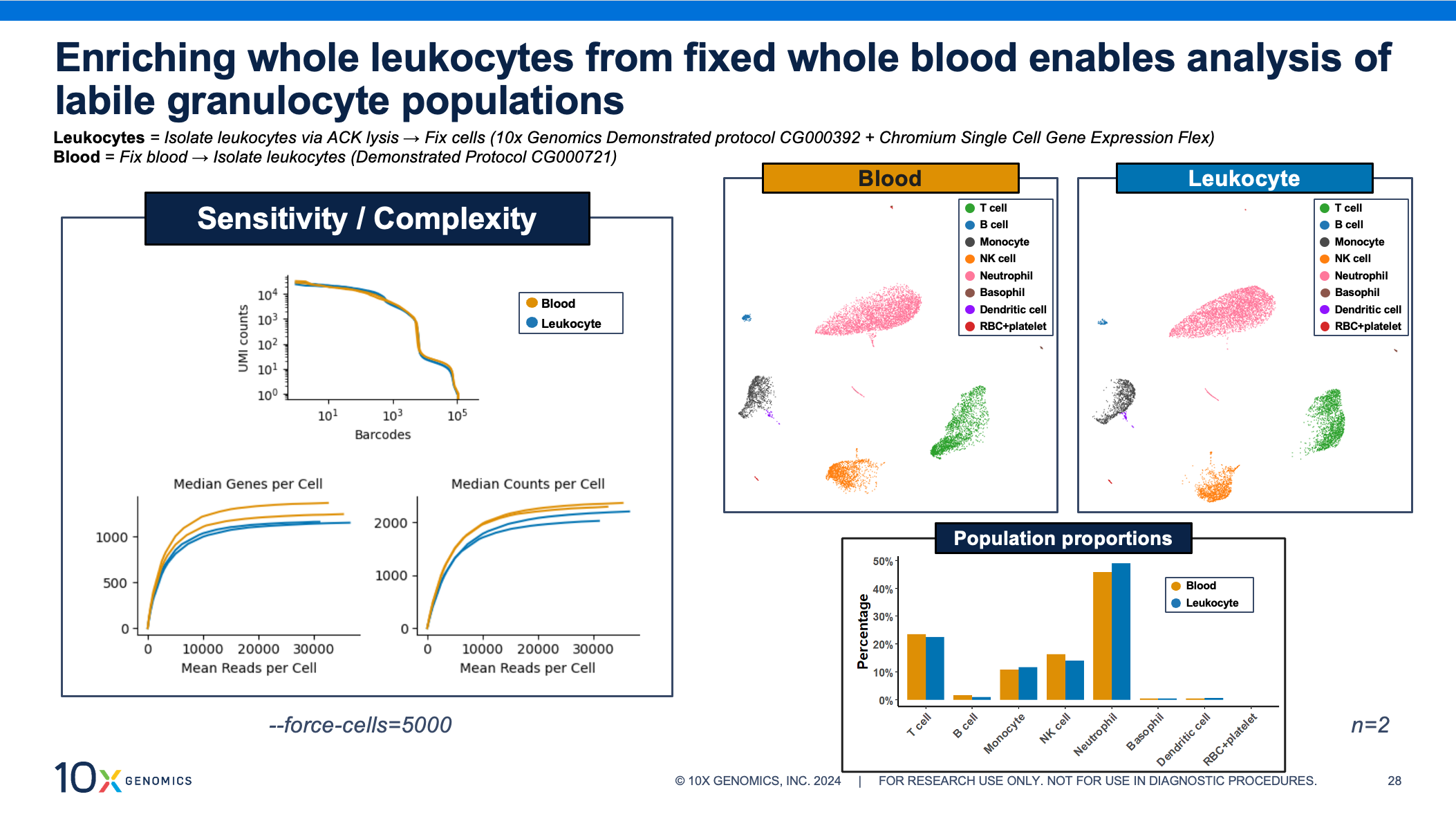
Granulocytes are key immune players but historically difficult to study via scRNA-seq. Fortunately, these fragile, low RNA–content cells see incredible performance with the whole blood fixation protocol. As with the PBMCs above, the data shows comparable transcriptomic complexity, sensitivity, cell clustering, and cell proportions, whether the leukocytes were enriched before or after fixation.
Longer storage options help streamline your research
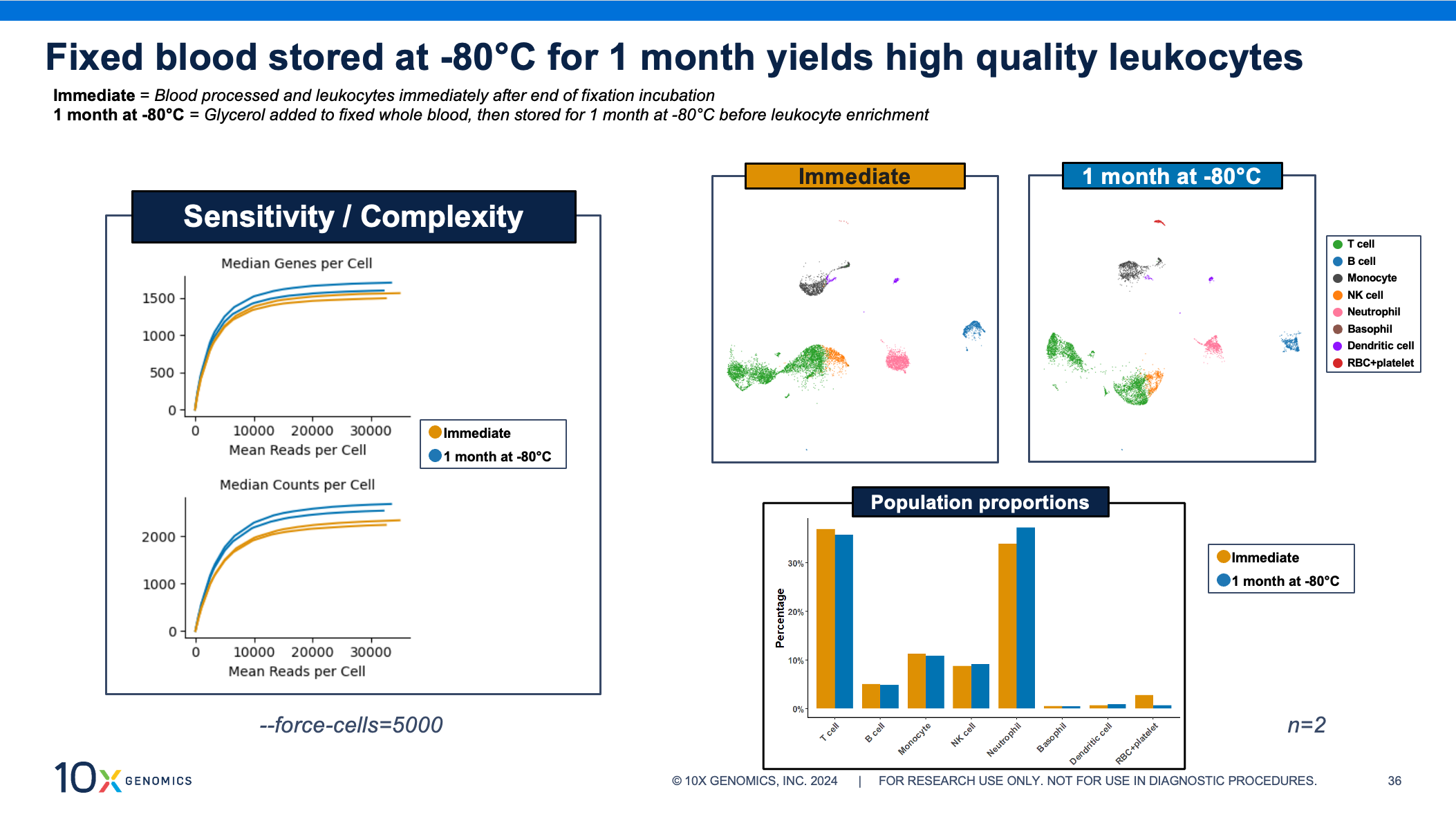
Now let’s turn to your longer storage option: adding glycerol to your fixed sample and storing it at –80°C for up to 1 month. That quick and easy step lets you choose when to thaw and isolate your cells of interest. Several samples can be thawed and batched together for processing through the rest of the single cell assay. Our scientists recommend working in batches of 8 samples, allowing you to move from sample fixation to cell isolation in just 2 or 3 hours, while the same number of samples processed individually could take up to 8 hours.
Even after 1 month of storage, these fixed whole blood samples yield high-quality single cell data. The sensitivity curves show similar transcriptome complexity, cell clustering, and population proportions between fixed blood processed and assayed immediately after fixation compared to fixed blood stored at –80°C.
Advice from our scientists: Q&A highlights
Getting the most out of your samples
What is the cell recovery efficiency with whole blood fixation?
From 1 mL of whole blood, fixed, you can get back a little over 1 million PBMCs or a little over 2 million total leukocytes. To put that into context, the minimum cell input for the Flex assay is 50K cells for hybridization, and you don’t lose very many cells during the washing steps in Flex. So just from 1 mL of blood you’re getting quite a large amount of cells for the downstream assay.
What is the percentage of cell loss observed after thawing the fixed sample? Any issue with cell clumping after thawing?
We don’t see any noticeable difference in the cell counts before and after freeze-thawing. And cell clumping is also not a big issue that we’ve seen.
Can you run the single cell assay without depleting red blood cells (RBCs)?
We strongly recommend enriching PBMCs or leukocytes. RBCs don’t have nuclei so they don’t really have much transcriptomic information, and that would kind of be a waste of your single cell runs. Since RBCs account for over 99% of the cell fraction in blood, if you load say 10,000 cells, only about 100 are going to be white blood cells. So you’re really losing a lot of information from your sample if you do not deplete the RBCs. Now, sometimes the cleanups aren’t perfect, and you’ll get some RBCs that get through the depletions, but that minor presence isn’t going to impact performance or consume a lot of sequencing reads at all. So I wouldn't worry about the minor presence of RBCs, but you should definitely be depleting them before you run our single cell kits.
Additional application compatibility
Is the fixed whole blood compatible with flow cytometry?
Fixed PBMCs and leukocytes can, in fact, be stained and analyzed with flow cytometry from the fixed whole blood. But you do need optimized antibody clones since we know that the formaldehyde fixation actually alters the epitopes and then that can affect the antibody binding efficiency. So yes, with the right panel that’s been optimized.
Did you know? Gene Expression Flex also provides highly sensitive whole transcriptome profiling starting from fresh, frozen, or fixed cells, nuclei, or whole tissue from human or mouse. You can learn more about single cell profiling with Flex by visiting the product page or this sample prep blog with advice from our scientists and a wealth of helpful resources for different sample types (including FFPE). Do you have specific questions on your tissue of interest? Check out our list of tested tissues for fresh/frozen and FFPE samples, provided by our technical support team.
This Q&A has been edited for clarity. Watch the full, on-demand webinar here.
References:
- Massoni-Badosa R, et al. Sampling time-dependent artifacts in single-cell genomics studies. Genome Biol 21: 112 (2020). doi: 10.1186/s13059-020-02032-0
- Qu HQ, et al. Single cell transcriptome analysis of peripheral blood mononuclear cells in freshly isolated versus stored blood samples. Genes (Basel) 14: 142 (2023). doi: 10.3390/genes14010142
- Barnes M, et al. Gene expression profiles from peripheral blood mononuclear cells are sensitive to short processing delays. Biopreserv Biobank 8: 153–162 (2010). doi: 10.1089/bio.2010.0009
- Guo C, et al. Single-cell transcriptomics reveal a unique memory-like NK cell subset that accumulates with ageing and correlates with disease severity in COVID-19. Genome Med 14: 46 (2022). doi: 10.1186/s13073-022-01049-3
- Ng MSF, et al. Deterministic reprogramming of neutrophils within tumors. Science 383: eadf6493 (2024). doi: 10.1126/science.adf6493
About the author:

Blog contributor: This article has been reviewed for scientific accuracy by Angela Churchill, PhD.

