A sequencing approach to T-cell receptor-antigen recognition
In this article, we discuss the value of T-cell receptor (TCR) sequencing for biological research, including recent bioinformatics developments that are helping researchers better manage and glean insights from the large amounts of data generated through single cell TCR-antigen mapping experiments. Read on to explore the latest research.
The importance of defining the “immunological synapse”
The true value of something can often be revealed in its absence. Imagine a scenario where T-cell antigen recognition was faulty—a condition known in the real world as “bubble boy” syndrome, or severe combined immunodeficiency. Immunity as we know it would be impossible. We wouldn’t be able to identify infectious agents when they enter our bodies or clear cells carrying viruses. There would be no immune memory formation to antigens that we’ve been exposed to before, making vaccination efforts futile. The very foundation of groundbreaking cancer immunotherapy approaches would be pulled out from under us. Our own cells would be at risk to dangerous autoimmune attacks. Painted in this light, the staggering reality is that T-cell receptor antigen–specific recognition is crucial for immune response mechanisms that keep us healthy. This makes specific, effective antigen recognition at the “immunological synapse,” that is, the molecular juncture between TCRs and the peptide-major histocompatibility complex (pMHC) on antigen presenting cells, the crux of the adaptive immune response (1).
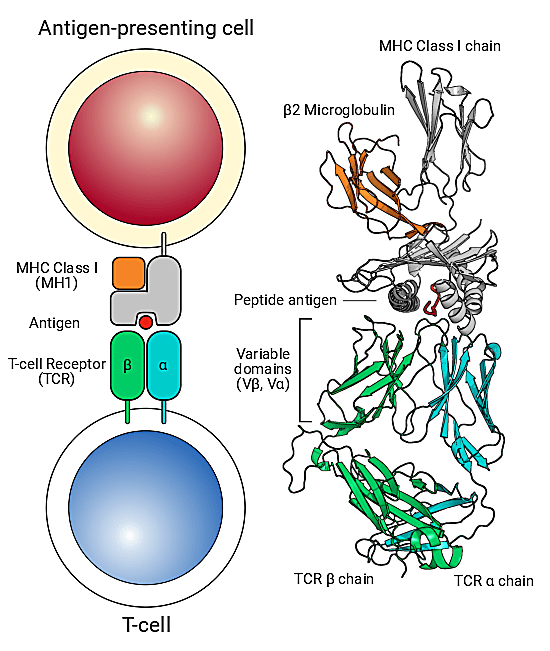
For these reasons, scientists are eager to catalog the huge diversity of antigen-specific T-cell receptors (TCR) roaming the immune system. New technology and data analysis platforms are allowing these efforts to scale like never before. The VDJdb database, an online resource to record TCR sequences and their cognate antigens, has dramatically expanded in recent years, with 61,049 records in 2019 compared to only 5,491 in 2017 (2). This includes an increase in paired alpha and beta TCR records obtained with the new capabilities enabled by high-throughput single cell sequencing. While progress has been made, there are many more records that must be generated to make inroads to the huge diversity of the immune repertoire, including categorization of unique antigen epitopes with known, specific TCRs—making this an epicenter of ongoing technological and bioinformatics innovation.
On the hunt for antigen specificity
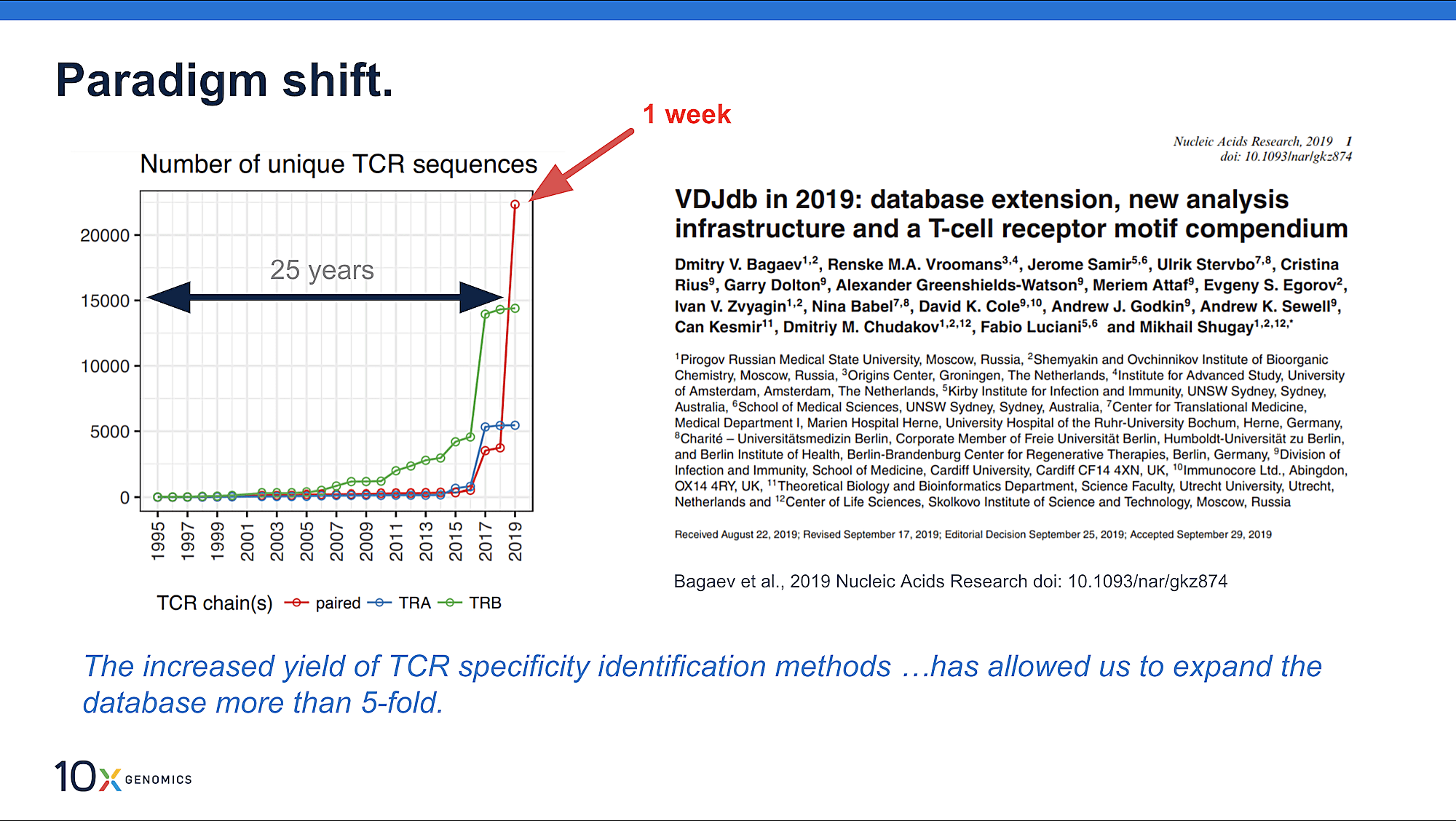
In 2019, a dataset leveraging 10x Genomics single cell immune profiling technology was contributed to the VDJdb database, adding 40 unique epitopes to the records, “more than any other high-throughput dataset produced so far” (2). This begs the question: what makes high-throughput single cell sequencing an effective solution for accurate TCR-specificity prediction? And what can improve this approach for even better results?
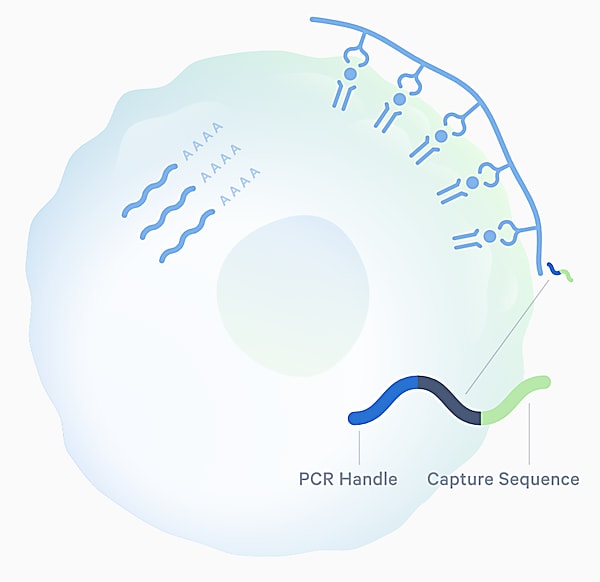
Access is the first hurdle single cell sequencing approaches can overcome, providing a readout of the sequences of TCRs and their cognate antigens at an efficiency and scale that matches the huge diversity of the TCR repertoire—estimated to range from 1015 to 1061 possible receptors in humans (3). The Chromium Single Cell Immune Profiling assay from 10x Genomics leverages pMHC multimers tagged with specialized oligonucleotides (Feature Barcode technology) to explore antigen specificity. When combined with single cell TCR sequencing, this allows for simultaneous capture of paired T-cell αβ-chain sequences and peptide–MHC multimers for a high resolution view of that binding interaction, as well as information about the identity and function of the T cell through gene expression analysis.
Many scientists are also looking for bioinformatics solutions to reduce this gap in data for TCR antigen specificity. Namely, the approach relies on TCR-pMHC binding data and learned TCR sequence features to predict antigen specificity; however, these are large datasets that often have a lot of complexity and nonspecific background noise, making it difficult to accurately validate TCR-pMHC–specific recognition (3). In their recent Science Advances publication, Zhang et al. described a data normalization approach called ICON (Integrative COntext-specific Normalization) that allowed them to identify reliable TCR-pMHC interactions from a 10x Genomics pMHC binding dataset by removing background noise. Assessing the binding profiles of CD8+ T cells taken from four healthy donors and tested across 44 dextramers, they were able to link 89% of 15,821 sequenced T cells with paired αβ chains to their antigen targets (3).
Zhang et al. also explored the use of neural networks to predict antigen specificity from TCR sequences, developing a model that can receive V(D)J genes and variable complementarity-determining regions (CDR) of the TCR as inputs. Others are actively exploring similar computational approaches, such as Montemurro et al. who shared in a recent Communications Biology publication, “The development of accurate prediction models is contingent on paired α/β TCR sequence data.” They noted the quality of the data with information for only one chain is lower than that of the data with paired CDRs. This was reflected in their results where models constructed from paired TCR data demonstrated significantly higher performance to similar models trained on data with CDR3β information only (4). Their results support the notion that both TCR chains contribute to TCR-antigen specificity and may have different relative importance to the binding interaction with pMHC. Importantly, this puts the requirement on single cell sequencing platforms to provide access to both chains as an essential factor for future discovery of accurate TCR-antigen–specific interactions.
For further reading about these bioinformatics solutions, explore the papers below:
- Zhang W, et al. A framework for highly multiplexed dextramer mapping and prediction of T cell receptor sequences to antigen specificity. Sci Adv 7: eabf5835 (2021). doi: 10.1126/sciadv.abf5835
- Montemurro A, et al. NetTCR-2.0 enables accurate prediction of TCR-peptide binding by using paired TCRα and β sequence data. Commun Biol 4: 1060 (2021). doi: 10.1038/s42003-021-02610-3
- Schattgen S, et al. Integrating T cell receptor sequences and transcriptional profiles by clonotype neighbor graph analysis (CoNGA). Nat Biotechnol (2021). doi: 10.1038/s41587-021-00989-2
Learn more about the latest immunology research powered by 10x Genomics solutions with on-demand presentations from our recent Global Infectious Disease Symposium. Watch here →
References:
- Cano LE and Lopera DE. Introduction to T and B lymphocytes. Autoimmunity: From Bench to Bedside [Internet]. Bogota (Colombia): El Rosario University Press (2013).
- Bagaev D, et al. VDJdb in 2019: database extension, new analysis infrastructure and a T-cell receptor motif compendium. Nucleic Acids Res 48: D1057–D1062 (2020). doi: 10.1093/nar/gkz874
- Zhang W, et al. A framework for highly multiplexed dextramer mapping and prediction of T cell receptor sequences to antigen specificity. Sci Adv 7: eabf5835 (2021). doi: 10.1126/sciadv.abf5835
- Montemurro A, et al. NetTCR-2.0 enables accurate prediction of TCR-peptide binding by using paired TCRα and β sequence data. Commun Biol 4: 1060 (2021). doi: 10.1038/s42003-021-02610-3
- Sudmeier L, et al. The CD8+ T cell landscape of human brain metastases. bioRxiv. Posted August 5, 2021. doi: 10.1101/2021.08.03.455000
