Note: 10x Genomics does not provide support for community-developed tools and makes no guarantees regarding their function or performance. Please contact tool developers with any questions. If you have feedback about Analysis Guides, please email analysis-guides@10xgenomics.com.
This article lists and describes some of the popular community-developed tools for analyzing 5' Single Cell Immune Profiling data after running Cell Ranger.
The Chromium 5' Single Cell Immune Profiling solution by 10x Genomics allows researchers to discover novel cell types, obtain a comprehensive immune repertoire, identify immune receptors mapped to antigens, and discover new biomarkers. Using Cell Ranger's efficient algorithms, Loupe Browser, and Loupe V(D)J Browser, researchers can detect rare V(D)J transcripts, identify cellular subpopulations, characterize paired TCR and BCR clonotypes, and identify antigen specific B and T cell clonotypes (with Barcode Enabled Antigen Mapping).
At the end of 2022, the scRNA-tools database (Zappia et al. 2018) contained about 1,399 software tools. Although only a small number of these tools (2.6%) are in the "Immune" category, they offer a variety of features and overlapping functionality that make it non-trivial for researchers to choose the right tool for single cell immunology data analysis in different biological contexts.
This article aims to help immunologists decide what to do after running Cell Ranger on 5' Single Cell Immune Profiling data. We describe various tools for immune profiling analysis. We also provide guidance for identifying tools necessary for analyzing and interpreting data downstream of Cell Ranger. To save time on Cell Ranger parameter optimization, it is important to select downstream tools that are best-suited to your research questions before running Cell Ranger.
A good starting point for assessing the accuracy and reliability of your data is the web_summary.html output by Cell Ranger multi or Cell Ranger vdj pipelines. A quick review of the web summary HTML file reveals any potential issues with data quality, enabling early intervention.
To help you get started, 10x Genomics provides a Knowledge Base article on interpreting web summary metrics for Single Cell Immune Profiling analysis.
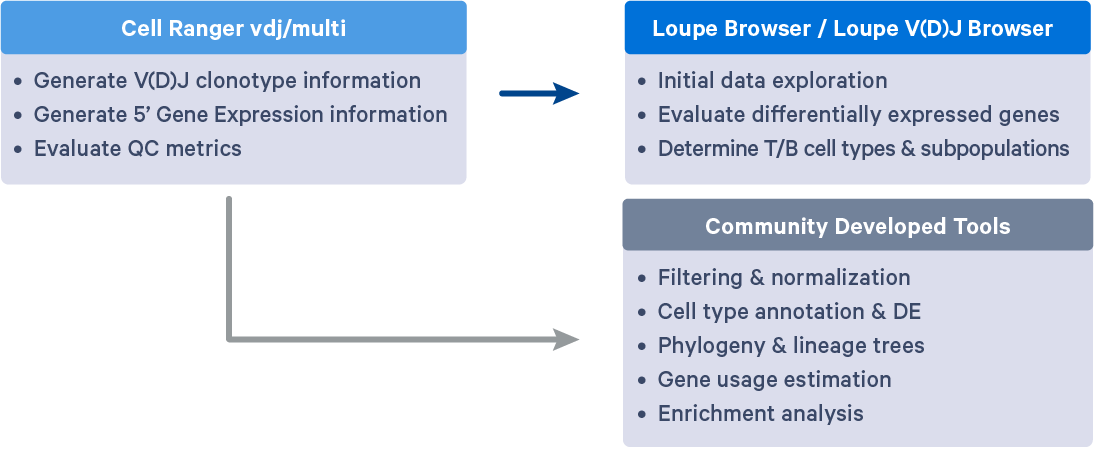
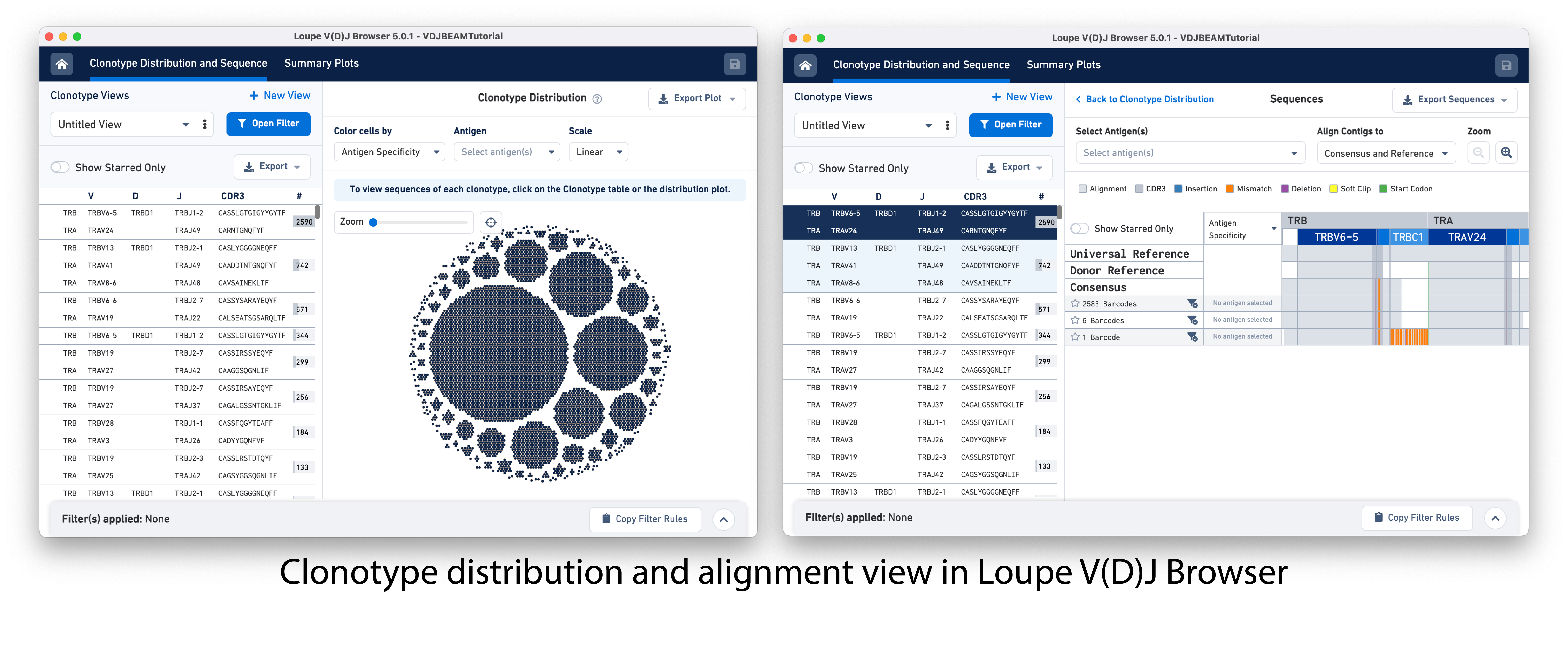
Cell Ranger outputs .cloupe and .vloupe files that can be input into Loupe Browser (LB) and Loupe V(D)J Browser (LVB), respectively. Visualizations available in LB and LVB can help further evaluate gene expression profiles, V(D)J gene usage, subpopulations of T/B cells, CDR3 sequence distribution, clonotype abundance and distribution, and even perform data QC or gene expression re-clustering. These initial analyses can provide insights into the diversity, specificity, and function of the Immune repertoire and ensure the reliability of downstream analyses.
Many community-developed tools are available for performing specialized analyses with Single Cell Immune Profiling data. Clonotype annotation, diversity evaluation, T/B-cell repertoire analysis, and other post-Cell Ranger steps are necessary for uncovering deeper biological insights related to repertoire diversity, clonal composition, and epitope specificity.
This table provides a high-level overview of a few popular community-developed tools, along with use cases for you to consider on your journey after running Cell Ranger:
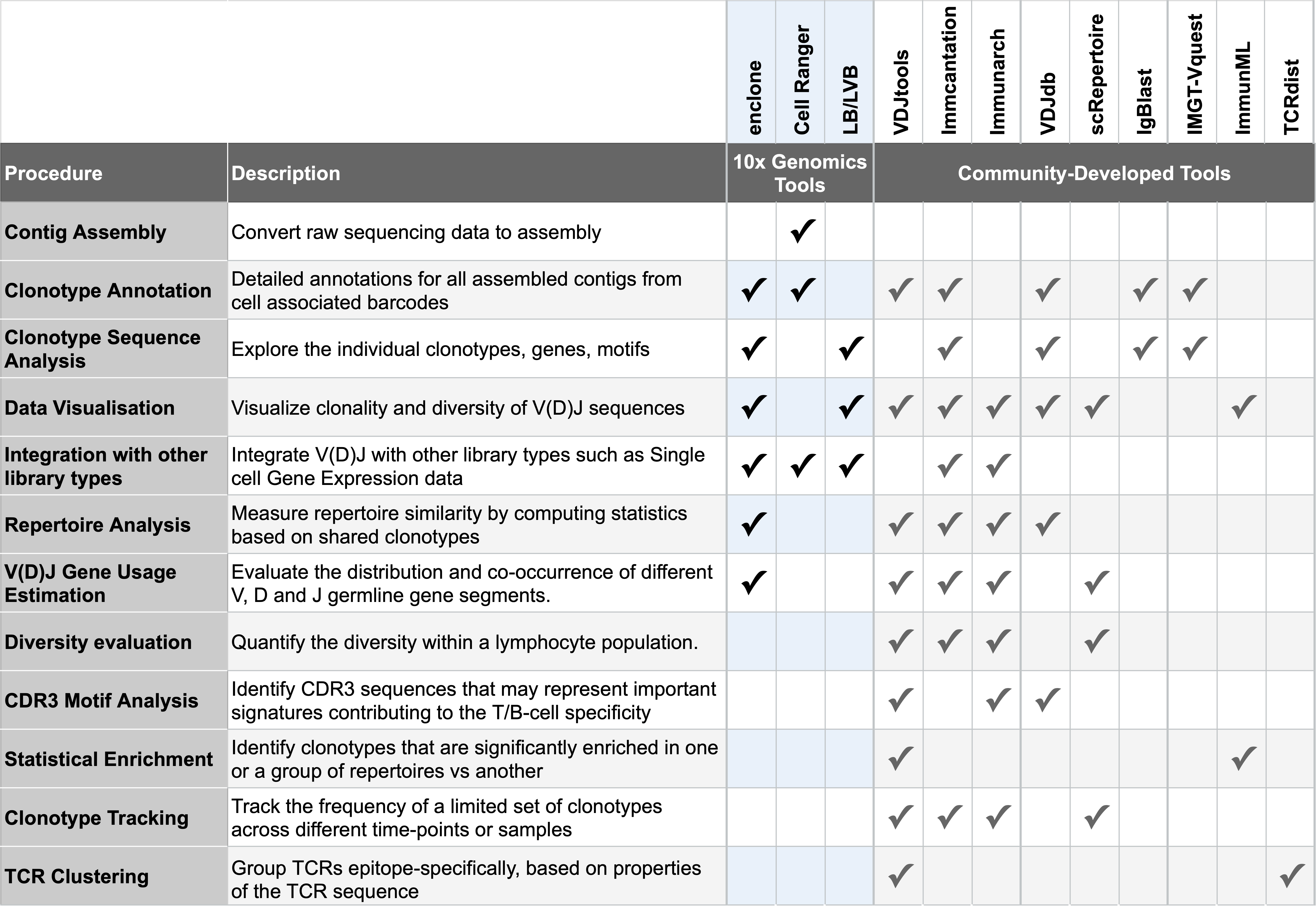
Clonotype annotation tools such as IMGT/V-QUEST, IgBLAST can be used to assign V(D)J gene usage, CDR3 sequences, and clonotype information to individual cells.
T or B cell repertoire analysis tools such as TCRdist and VDJdb can be used to identify shared clonotypes across multiple samples and explore the specificity of an immune response.
VDJtools, scRepertoire, and Immunarch can be used to analyze and visualize gene rearrangements to study clonal diversity, gene usage, and junction properties of the rearrangements. These tools allow you to compare gene rearrangements across samples and between cell types.
The software package Immcantation (in the programming language R) can be used to define clonal groups and build lineage trees. Many tutorials are available to get you started with Immcantation. Specifically, the tutorial on defining B cell clonal families and building B cell lineage trees uses 10x Genomics BCR sequencing data.
Some platforms like ImmuneDB (not listed in the table) also provide a user-friendly web framework that allows for storage in addition to tools for analyzing clonal composition, diversity, and immune repertoire evolution.
These are a small subset of the many community-developed tools available for analyzing and interpreting single cell immunology data. Depending on the specific research question and dataset, other tools may be more appropriate. It is important to carefully evaluate the strengths and limitations of each tool before use.
- Zappia L, Phipson B, & Oshlack A. Exploring the single-cell RNA-seq analysis landscape with the scRNA-tools database. PLOS Computational Biology 14:6, 2018.
- Gadala-Maria D, et al. Identification of Subject-Specific Immunoglobulin Alleles From Expressed Repertoire Sequencing Data, Frontiers in Immunology 10:129, 2019.
- Arunkumar M & Zielinski. T-Cell Receptor Repertoire Analysis with Computational Tools—An Immunologist’s Perspective. Cells 10:12, 2021.
- Nazarov V, et al. Immunarch: Bioinformatics Analysis of T-Cell and B-Cell Immune Repertoires, 2022.
- Valkiers S, et al. Recent advances in T-cell receptor repertoire analysis: Bridging the gap with multimodal single-cell RNA sequencing. ImmunoInformatics 100009, 2022.
- Vander H, et al. pRESTO: A Toolkit For Processing High-throughput Sequencing Raw Reads of Lymphocyte Receptor Repertoires. Bioinformatics 30:1930-2, 2014.