Functional genomics at single cell resolution
Link CRISPR edits to cell phenotype
Directly link CRISPR edits and gene expression phenotypes at single cell resolution.
Gain deeper insights
Access comprehensive insights, beyond cell fitness, with single cell multiomic profiling.
Scale your CRISPR screens
Analyze genetic perturbation effects across hundreds of genes in hundreds of thousands of cells.
Reduce time to results
Quicker time to gene expression results, compared to lengthy culture times for cell survival assays.
Single cell resolution
Interrogate disease pathway and perturbation effects at scale, cell by cell.
Streamline data analysis
Explore gene expression profiles from perturbations with easy-to-use software.
Explore what you can do
Scalable functional genomics
Validate hits identified in GWAS and QTL studies
Mechanistic insights into disease biology
Reveal genetic contributions to disease across the entire genome
Videos
Resources
Publications
Nat Biotechnol. 2020, Replogle JM, et al.
Nat Commun. 2021, Yang L, et al.
Sci Adv. 2021, Lopes R et al.
Our end-to-end solution
Chromium instrument with Next GEM technology
Chromium Single Cell reagents
With our reagent kits, explore cellular diversity through gene expression, cell surface proteins, CRISPR perturbations, and more.

Analysis and visualization software
World-class technical and customer support
Our expert support team can be contacted by phone or email.

1. Design your CRISPR library
Pick the target genes whose function you’d like to further investigate—from a deep dive into a focused set of tens of genes to a broad screen of hundreds of genes that comprise many pathways. Then, design your sgRNAs to order, with one of many freely available CRISPR guide RNA design tools.
2. Prepare your sample
Transduce and stimulate your cells—infect your cells, select the cells expressing your CRISPR guide, and apply your stimulus. Harvest your cells, and start exploring across hundreds of thousands of cells. Follow best practices for washing, counting, and concentrating cells to minimize the presence of cellular aggregates, and retrieve high-quality single cell suspensions.
3. Construct your 10x library
Construct a 10x barcoded library using our reagent kits and a compatible Chromium instrument. Each member of the Chromium instrument family encapsulates each cell with a 10x barcoded Gel Bead in a single partition. Within each nanoliter-scale partition, cells undergo reverse transcription to generate cDNA for both mRNA and CRISPR guides, each of which shares a 10x Barcode with all cDNA from its individual cell of origin.
4. Sequence
Sequence the resulting 10x barcoded library on compatible standard NGS short-read sequencers for massive transcriptional profiling of hundreds of thousands of individual cells.
5. Analyze your data
Use our Cell Ranger analysis software to automatically assign guides on a per cell basis and directly assess perturbation effects of a guide on the target gene as well as the entire transcriptome.
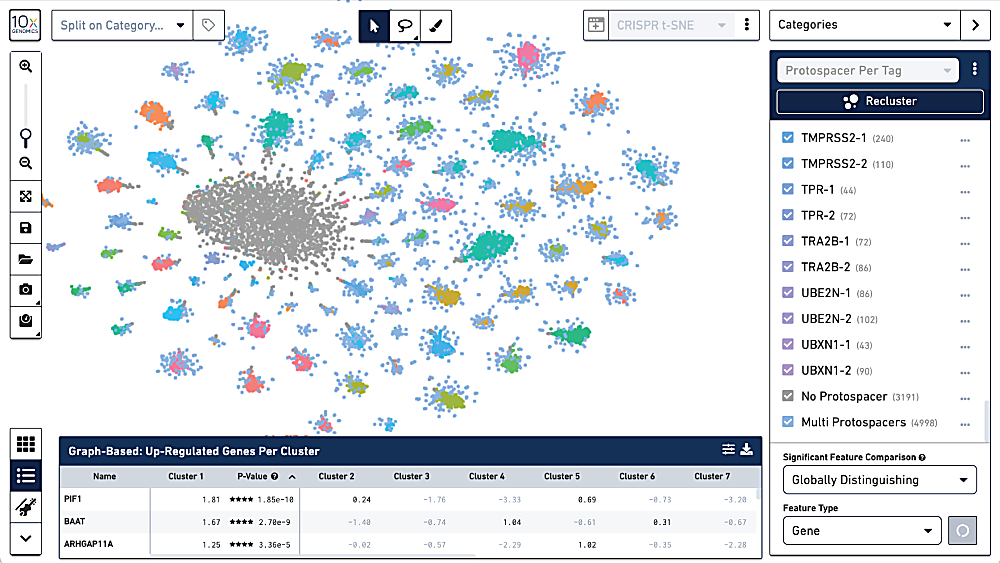
6. Visualize your data
Interactively explore your results with our Loupe Browser visualization software. Study expression patterns for genes of interest as well as resulting perturbation phenotypes across guide targets, and perform comparative analysis between clusters and samples.
Frequently Asked Questions
How do single cell CRISPR screens differ from bulk CRISPR screens?
Single cell CRISPR screens from 10x Genomics capture and barcode CRISPR sgRNAs and cellular transcripts within each cell, providing a direct link between perturbation and phenotype in a pooled guide library format. Additionally, the readout provides information for each single cell, enabling the resolution of clusters and subpopulations of cells within a sample.
What types of CRISPR perturbations have been demonstrated with 10x Genomics technology?
CRISPR knockout, inhibition, and activation have all been demonstrated on 10x Genomics single cell technologies.
What is the difference between the 10x Genomics Single Cell 5' and 3' CRISPR Screening assays, and which should I use?
Our 5’ CRISPR screening assay is compatible with most pre-existing Cas9 guide RNA vectors. If you have an existing CRISPR library or are starting a single cell CRISPR screen for the first time, we recommend using our Single Cell 5’ v2 assay which is compatible with most guide libraries with little to no optimization required.
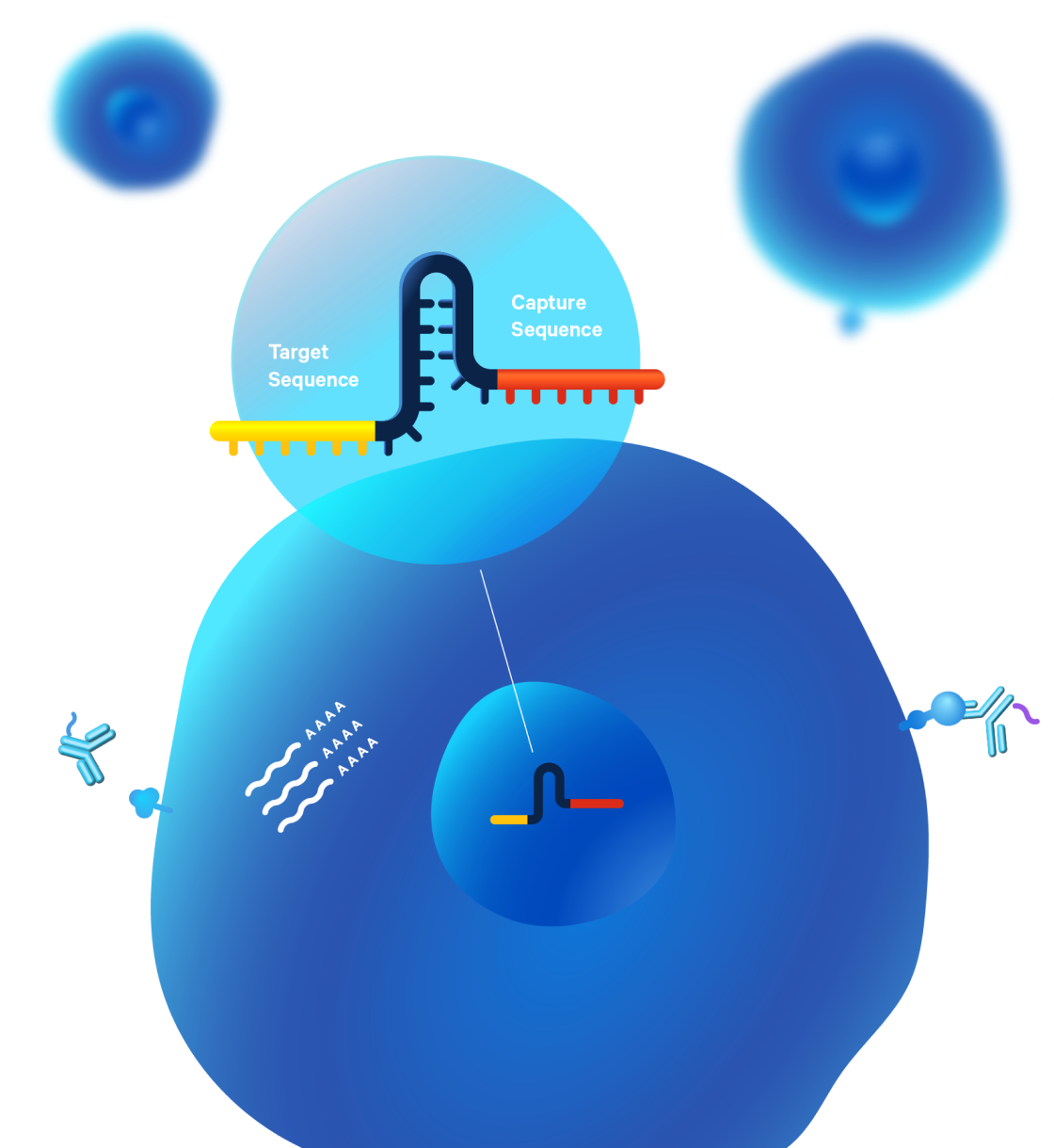
Our 3’ CRISPR screening assay requires that a specific Capture Sequence be inserted into the vector at the appropriate location for capture of the guide. If you want to compare your single cell CRISPR experiment to data from previous experiments using 3’ single cell gene expression with Feature Barcode technology, we recommend continuing to use our Single Cell 3’ v3.1 assay to minimize batch effects.
Where can I find guide vectors compatible with Single Cell 3’ CRISPR Screening?
Our compatible product partner, SigmaAldrich®*, can design and manufacture custom lentiviral Feature Barcode compatible guide libraries. Feature Barcode enabled CRISPR guide vectors can also be found on Addgene, pBA900 and pBA904 deposited by the Weissman Lab.
SigmaAldrich® 10x Genomics Custom CRISPR Lentiviral Products
Single guide expression CRISPR vectors
Dual guide expression CRISPR vector
*Sigma-Aldrich is a trademark of Merck KGaA, Darmstadt, Germany or its affiliates. All other trademarks are the property of their respective owners. Detailed information on trademarks is available via publicly accessible resources.
Where can I find resources to help with CRISPR guide design?
Gene-editing or functional genomics cores will often have resources or can help guide CRISPR guide design. Addgene also provides a guide to choosing the best CRISPR sgRNA design tool, or you can provide MilliporeSigma with your target gene list and they will design the guides for the library pool you order.