The pipeline output directory, described in Understanding Output, contains all of the data produced by one invocation of a pipeline (a pipestance) as well as rich metadata describing the characteristics of each stage. This directory contains a specific structure that is used by the Martian pipeline framework to track the state of the pipeline as execution proceeds.
Cell Ranger's notion of a pipeline is very flexible in that a pipeline can be composed of stages that run stage code or sub-pipelines that may themselves contain stages or sub-pipelines.
Cell Ranger pipelines follow the convention that stages are named with verbs (e.g., ALIGN_READS, MARK_DUPLICATES, FILTER_BARCODES) and sub-pipelines are named with nouns and prefixed with an underscore (e.g., _BCSORTER). Each stage runs in its own directory bearing its name, and each stage's directory is contained within its parent pipeline's directory.
For example, the cellranger-arc mkfastq pipeline has the following process graph:
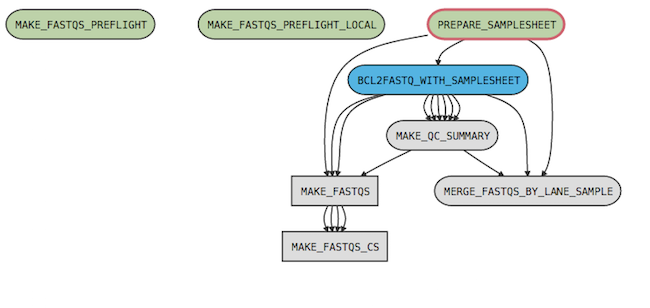
where
MAKE_FASTQS_CSis the top-level pipeline stageMAKE_FASTQSis a sub-pipeline contained inMAKE_FASTQS_CSPREPARE_SAMPLESHEET,BCL2FASTQ_WITH_SAMPLESHEET,MAKE_QC_SUMMARY, andMERGE_FASTQS_BY_LANE_SAMPLEare stages contained in theMAKE_FASTQSsub-pipeline.MAKE_FASTQS_PREFLIGHTandMAKE_FASTQS_PREFLIGHT_LOCALare preflight stages, which validate inputs prior to running the other stages. These also belong toMAKE_FASTQS, but have no connections to other stages because they don't produce any outputs.
The MAKE_FASTQS_CS stage is not strictly necessary since it contains no stages and only one child pipeline (MAKE_FASTQS); however, it serves to mask some of the low-level inputs required by the MAKE_FASTQS pipeline.
Every pipestance operates wholly inside of its pipeline output directory. When the pipestance completes, this pipestance output directory contains three outputs: metadata files, the pipestance output file directory, and the top-level pipeline stage directory.
- Metadata files are files prefixed with an underscore (
_) and usually contain unstructured text or JSON-encoded arrays and hashes. - The pipestance output file directory is a directory called
outs/that contains the pipestance's output files. - The top-level pipeline stage directory is a directory named according to the top-level pipeline stage that contains the child stage directories that compose this pipestance.
The top-level pipeline stage directory is a stage directory that contains any number of child stage directories as well as one stage output directory for each fork run by that stage. The top-level pipeline stages for Cell Ranger ARC are:
MAKE_FASTQS_CSfor cellranger-arc mkfastqSC_ATAC_GEX_COUNTER_CSfor cellranger-arc count
Most of the Cell Ranger ARC pipelines contain single-fork stages, which means there is one fork0 stage output directory within each stage directory. Chunk output directories are a subset of stage output directories that additionally contain runtime information specific to the job or process being run by that chunk (e.g., a process ID or cluster job ID).
For example, the cellranger-arc mkfastq pipeline's pipeline output directory contains the following directory structure:
| _log | Metadata file |
| outs/ | Pipestance output file directory |
| MAKE_FASTQS_CS/ | Top-level pipeline stage directory |
| MAKE_FASTQS_CS/fork0/ | Stage output directory |
| MAKE_FASTQS_CS/fork0/files/ | Stage output files |
| MAKE_FASTQS_CS/MAKE_FASTQS/ | Stage directory |
| MAKE_FASTQS_CS/MAKE_FASTQS/fork0/ | Stage output directory |
| MAKE_FASTQS_CS/MAKE_FASTQS/fork0/files/ | Stage output files |
| MAKE_FASTQS_CS/MAKE_FASTQS/BCL2FASTQ_WITH_SAMPLESHEET/ | Stage directory |
| MAKE_FASTQS_CS/MAKE_FASTQS/BCL2FASTQ_WITH_SAMPLESHEET/fork0/ | Stage output directory |
| MAKE_FASTQS_CS/MAKE_FASTQS/BCL2FASTQ_WITH_SAMPLESHEET/fork0/chnk0/ | Chunk output directory |
The metadata contained in the pipeline output directory includes
| File Name | Description |
|---|---|
| Metadata cache that is populated when a pipestance completes to minimize re-aggregation of metadata | |
| The MRO call used to invoke this pipestance | |
The log messages that are reported to your terminal window when running cellranger-arc commands | |
_mrosource | The entire MRO describing the pipeline with all @include statements dereferenced |
_perf | Detailed runtime performance data for every stage in the pipestance |
_timestamp | The start and finish time for this pipestance |
_vdrkill | A list of all of the volatile data (temporary files) removed during pipeline execution as well as total number of files and bytes deleted |
_versions | Versions of the components used by the pipeline |
Stage directories contain stage output directories, stage output files, and the stage directories of any child stages or pipelines.
Stage output directories typically contain:
| File Name | Contents |
|---|---|
files/ | Directory containing any files created by this stage that were not considered volatile (temporary) |
split/ | A special stage output directory for the step that divided this stage's input into parallel chunks |
chnkN/ | A chunk output directory for the Nth parallel chunk executed |
join/ | A special stage output directory for the step that recombined this stage's parallel output chunks into a single output dataset again |
_complete | A file that, when present, signifies that this stage has successfully completed |
_errors | A file that, when present, signifies that this stage failed. Contains the errors that resulted in stage failure. |
_invocation | The MRO call used to execute this stage by the Martian framework |
_outs | The output files generated by this stage |
_vdrkill | A list of all of the volatile data (temporary files) removed during pipeline execution as well as total number of files and bytes deleted |
Chunk output directories are a subset of stage output directories that, in addition to the aforementioned stage output, may contain:
| File Name | Contents |
|---|---|
_args | The arguments passed to the stage's stage code |
_jobinfo | Metadata describing the stage's execution, including performance metrics, job manager jobid and jobname, and process ID |
_jobscript | The script submitted to the cluster job manager (cluster mode |
_stdout | Any stage code output that was printed to the stdout stream |
_stderr | Any stage code output that was printed to the stderr stream |
These metadata files should be treated as read-only, and altering the contents of metadata files is not recommended.