Mapping the second brain: Single cell characterization of the enteric nervous system reveals clues to IBD
April is Irritable Bowel Syndrome (IBS) Awareness Month, and chances are you know someone who struggles with this common disorder. Though it’s estimated that 10–15% of the population has IBS—or between 25 and 45 million people in the United States—it is commonly undiagnosed (1). Worse, the exact cause of IBS remains unknown, and while treatments do exist, they may not completely address symptoms.
Inflammatory bowel disease (IBD), which is diagnosed by the presence of inflammation in the gut, is an umbrella term that encompasses Crohn’s disease, ulcerative colitis, and other syndromes, like IBS. Because IBD and IBS are believed to involve the gut–brain axis, and this enteric nervous system has not yet been well characterized, single cell tools will help enable a deeper understanding of both the heterogeneity of this “second brain” and its complexity, or, how these cells interact with surrounding endothelial, stromal, and immune cells. In this post, we take a look at one research paper that uses single cell sequencing to build an atlas of the human and mouse colon in an effort to understand the disease mechanism behind IBD—and how this knowledge might inform the development of improved therapies.
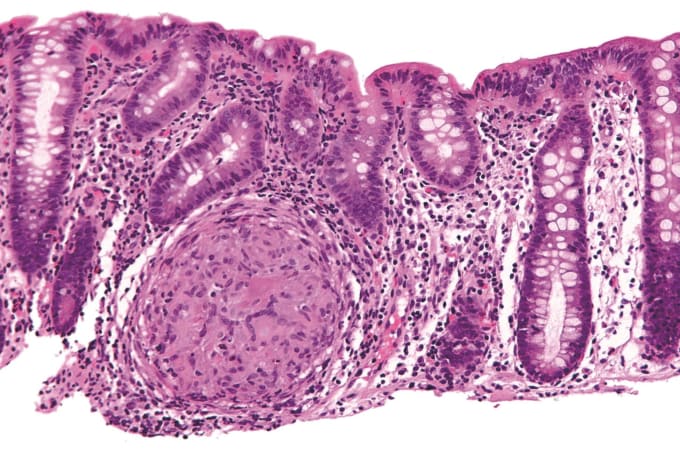
Inflammatory bowel disease (IBD) is a term used to describe an autoimmune disease that is characterized by chronic inflammation of the gastrointestinal tract, two types of which are Crohn's disease and ulcerative colitis. People with Crohn's and ulcerative colitis may have abdominal pain, diarrhea, bleeding from the rectum, pelvic cramps or muscle spasms, and weight loss leading to anemia. Potentially caused by IBD, irritable bowel syndrome (IBS) is diagnosed by an array of symptoms that affect bowel movements, including cramping, abdominal pain, bloating, gas, and diarrhea or constipation, or both.
While inflammatory bowel diseases are considered autoimmune diseases, the causes of IBD and related disorders like IBS are not known. In IBD, factors leading to inflammation in the intestine can include diet, changes in the gut microbiome, and loss of integrity of the intestinal epithelium—which allows for altered communication between the immune system and the gut. In IBS, factors that appear to play a role include similar things, such as stronger or weaker than normal muscle contractions in the intestine, infection, early life stress, and changes in gut microbes.
Even though the root causes are still unknown, scientists believe that the enteric nervous system is involved. To start, the entire nervous system is composed of the central nervous system, which consists of the brain and spinal cord, and the peripheral nervous system. In turn, the peripheral nervous system consists of the somatic nervous system, which acts on voluntary movement of skeletal muscles, and the autonomic nervous system, which acts involuntarily (or, unconsciously on our part) on smooth muscle and glands to regulate bodily functions. Even further down the line, the autonomic nervous system comprises the sympathetic nervous system, the parasympathetic nervous system, and the enteric nervous system (ENS).
The ENS is made up of neurons localized to the gut that communicate via neurotransmitters similar to those found in the brain that control gastrointestinal function, including intestinal motility, digestion, and barrier defense. Known as the “second brain,” recent studies show that the ENS is complex and heterogeneous—and that a range of both intestinal and neurological disorders associated with gut dysfunction, such as autism spectrum disorders and Parkinson’s disease, are tied back to faulty function of the ENS.
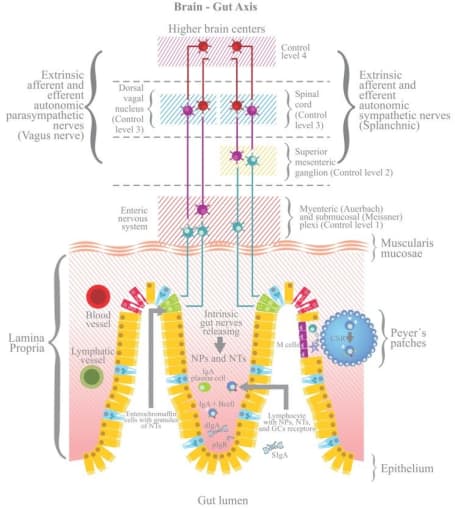
Opening the aperture: Seeing beyond neurons
Because the ENS consists of a number of rare, complex cell types, it has up until now been difficult to fully characterize the “second brain.” In a recent paper published in Cell (2), researchers led by Eugene Drokhlyansky, PhD, in the lab of Aviv Regev, PhD, at the Broad Institute of MIT and Harvard, sought to profile the ENS of the adult mouse and human at single cell resolution with the hope of exploring how neuronal diversity may contribute to interactions with other cell types and lead to inflammatory and other conditions. To generate a map of the adult ENS at single cell resolution, they developed two methods for single-nucleus RNA-seq (snRNA-seq) using 10x Genomics Chromium Single Cell Gene Expression: “ribosomes and intact single nucleus” (RAISIN) RNA-seq for isolation of intact nuclei and ribosome-bound RNA—a modified method for optimal nucleus extraction that retains the nuclear envelope and ribosomes on the outer nuclear membrane—and “mining rare cells sequencing” (MIRACL-seq) for label-free profiling of rare cell types, such as enteric neurons—a method for profiling rare cell types by overloading droplets with single nuclei/cells.
Using RAISIN RNA-seq to profile 2,657 neurons and 3,039 glia from the colons of adult mice, they identified 21 neuronal and three glial cell subsets. (From this, they estimate that neurons comprise less than 1% of all nuclei in the mouse colon.) The neuronal subsets include types of motor neurons, sensory neurons, interneurons (which relay signals between neurons), and secretomotor/vasodilator neurons (which trigger secretions and fluid movement). One subset of sensory neurons express brain-derived neurotrophic factor, which is elevated in patients with IBS (3). They also found that different cells’ gene expression is impacted by several factors—location in the intestine, circadian phase, and age—and that neurons from the distal colon have higher expression of many neurotransmitter receptors.
For the second part of the experiment, they looked to characterize the ENS of non-transgenic mouse colon, mouse ileum, and human colon. They profiled 704,314 mouse colon nuclei by MIRACL-seq and found a diversity of cell types, including 18 subsets of neurons and three subsets of glia; then, they profiled 483,221 mouse ileum nuclei, also finding neurons (12 subsets), glia (two subsets), and other rare cell types, including interstitial cells of Cajal (ICCs). To construct an ENS atlas of the human colon, they used MIRACL-seq to profile 436,202 nuclei from cancer-adjacent regions of colon resections from 16 colorectal cancer patients. Analysis revealed diverse cell types—14 subsets of neurons, as well as glia, adipocytes, endothelial cells, fibroblasts, immune cells, ICCs, and myocytes.
Comparing the 14 subsets of human colon neurons to the 18 subsets from the mouse, they discovered that there are conserved transcriptional programs for each neuron type between species but that there were expression differences in the melanocortin, leptin, and serotonin pathways, possibly reflecting the different feeding behaviors of the two species. Finding conserved gene expression across species highlights the importance of mouse research models in studying human diseases.
To examine interactions between the ENS and other colon cell types, they mapped thousands of receptor–ligand pairs onto their data. They found interactions between the ENS and parenchymal, stromal, and immune cell types, potentially reflecting the ability of the ENS to interact with the immune environment.
Finally, they asked whether enteric neurons expressed risk genes for diseases, including IBD. After mapping 185 disease risk genes onto cells from the human colon mucosa and muscularis, they found that most IBD risk genes were enriched in mucosal cells (the mucosa is the lining of the colon). However, they saw enrichment of several IBD risk genes in enteric neurons, including BTBD8, GRP, JAZF1, NDFIP1, PLA2R1, and CNTNAP2.
Clearing the path for research into diseases along the gut–brain axis
Up until now, the ENS was poorly characterized, in general, making it difficult to create research models of ENS-related diseases in mice. Building a comprehensive cell atlas of the enteric nervous system, which includes rare and hard-to-isolate cells, not only lends insight into the building blocks of the “second brain,” but also into the interaction of the ENS with different biological systems, including the gut, immune, and nervous systems. With this global reference in hand, scientists can now further study healthy ENS function as well as how dysfunction may lead to altered interactions with the immune system and brain, contributing to a diversity of diseases involving systemic inflammation.
References:
- https://aboutibs.org/what-is-ibs/facts-about-ibs/
- Drokhlyansky E, et al. The human and mouse enteric nervous system at single-cell resolution. Cell 182: 1606–1622.e23, 2020.
- Yu Y-B, et al. Brain-derived neurotrophic factor contributes to abdominal pain in irritable bowel syndrome. Gut 61: 685–694, 2012.
