Demonstrated Protocol for Nuclei Isolation for Single Cell ATAC Solution
The eukaryotic genome is highly packaged to fit into the very limited nuclear space. As a result, access to genomic information is highly regulated. The accessibility of regions within the genome reveals a great deal about the state of the cell. ATAC-seq, or Assay for Transposase-Accessible Chromatin coupled with next-generation sequencing, is a technique to locate accessible chromatin regions.
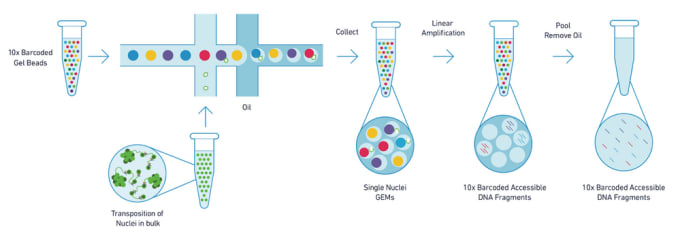
ATAC-seq is based on the use of a transposase enzyme, that allows for highly efficient cutting of exposed chromatin DNA and simultaneous ligation of specific sequences, called adapters. Adapter-ligated DNA fragments are then isolated, amplified by PCR and used for next generation sequencing. Sequencing reads are used to infer regions of increased accessibility (open chromatin), as well as to map regions of transcription factor binding and nucleosome positioning.
Current methods for assaying chromatin structure and composition often require a large number of cells as input material, averaging out heterogeneity in cellular populations. In many cases, rare and important cellular subtypes cannot be acquired in amounts sufficient for genome-wide chromatin analyses. To address these shortcomings, 10x Genomics has launched its Chromium Single Cell ATAC Solution. With as little as a few hundred cells, the Single Cell ATAC Solution allows profiling of chromatin structure at the single cell level, providing an unprecedented view of the heterogeneity of chromatin regulation.
A well-known problem regarding ATAC-seq datasets, is that they usually contains a large percentage of reads from mitochondrial DNA. Large amounts of mitochondrial contamination result in higher wasted data and more sequencing dollars for customers. The 10x Single Cell ATAC workflow has been optimized for low mitochondrial reads from samples. A key step to ensure this, is to carefully extract nuclei from cells, while ensuring the mitochondria stays intact. The combination of lysis detergents in the 10x developed workflow is critical for the low mitochondrial reads. Thus, the Nuclei Isolation for Single Cell ATAC Sequencing Demonstrated Protocol (CG000169) is an integral part of the product offering to ensure low mitochondrial reads and high quality data.
This demonstrated protocol outlines processes for isolating nuclei from both fresh and cryopreserved cell lines and as well as more fragile primary cells. As many samples may have limiting sources and material, we have also provided guidance for low input samples. The protocol has been validated on PBMCs as well as GM12878 and EL4 cells. With further optimization, the demonstrated protocol may be applicable to most cell types. It is important to note that this protocol is distinct from the Isolation of Nuclei for Single Cell RNA Sequencing (CG000124), by the fact that nuclei prepared for Single Cell ATAC profiling will require a permeabilized nuclear membrane for diffusion of transposition reagents. For this reason, users should make sure to use the proper demonstrated protocol to ensure assay performance. Note that not all demonstrated protocols on the 10x Support website are compatible with the Single Cell ATAC assay.
To streamline sample preparation and reduce variability, the Chromium Single Cell ATAC Solution Library Kit will include Nuclei Buffer. This component is provided as a concentrated stock solution, and a diluted working stock must be used for resuspending nuclei in the last step of the Nuclei Isolation for Single Cell ATAC Sequencing Demonstrated Protocol (CG000169). The use of the diluted Nuclei Buffer for nuclei suspension is critical for optimal Single Cell ATAC assay performance. The composition of the Tris-based diluted Nuclei Buffer, including Magnesium concentration, has been optimized for the Transposition and Barcoding steps in the Single Cell ATAC protocol. Suspension of nuclei in a different buffer may not be compatible with these protocol steps.
We are excited to release the Chromium Single Cell ATAC Solution, and hope you’ll join us as we continue to push the boundaries of single-cell technologies!
Feel free to contact us at support@10xgenomics.com with your questions regarding this, or other solutions.
